Example : k times repetition with the list of k input files¶
DeepBiome package takes microbiome abundance data as input and uses the phylogenetic taxonomy to guide the decision of the optimal number of layers and neurons in the deep learning architecture.
To use DeepBiome, you can experiment (1) k times repetition or (2) k fold cross-validation. For each experiment, we asuume that the dataset is given by - A list of k input files for k times repetition. - One input file for k fold cross-validation.
This notebook contains an example of (1) k times repetition for the deep neural netowrk using deepbiome.
1. Load library¶
First, we load the DeepBiome package. The DeepBiome package is built on the tensorflow and keras library
[1]:
import os
import logging
import json
from pkg_resources import resource_filename
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
pd.set_option('display.float_format', lambda x: '%.03f' % x)
np.set_printoptions(formatter={'float_kind':lambda x: '%.03f' % x})
from deepbiome import deepbiome
Using TensorFlow backend.
2. Prepare the dataset¶
In this example, we assume that we have a list of k input files for k times repetition.
DeepBiome needs 4 data files as follows: 1. the tree information 1. the lists of the input files (each file has all sample’s information for one repetition) 1. the list of the names of input files 1. y
In addition, we can set the training index for each repetition. If we set the index file, DeepBiome builds the training set for each repetition based on each column of the index file. If not, DeepBiome will generate the index file locally.
Each data should have the csv format as follow:
Example of the tree information¶
First we need a file about the phylogenetic tree information. This tree information file should have the format below:
[2]:
tree_information = pd.read_csv(resource_filename('deepbiome', 'tests/data/genus48_dic.csv'))
tree_information
[2]:
| Genus | Family | Order | Class | Phylum | |
|---|---|---|---|---|---|
| 0 | Streptococcus | Streptococcaceae | Lactobacillales | Bacilli | Firmicutes |
| 1 | Tropheryma | Cellulomonadaceae | Actinomycetales | Actinobacteria | Actinobacteria |
| 2 | Veillonella | Veillonellaceae | Selenomonadales | Negativicutes | Firmicutes |
| 3 | Actinomyces | Actinomycetaceae | Actinomycetales | Actinobacteria | Actinobacteria |
| 4 | Flavobacterium | Flavobacteriaceae | Flavobacteriales | Flavobacteria | Bacteroidetes |
| 5 | Prevotella | Prevotellaceae | Bacteroidales | Bacteroidia | Bacteroidetes |
| 6 | Porphyromonas | Porphyromonadaceae | Bacteroidales | Bacteroidia | Bacteroidetes |
| 7 | Parvimonas | Clostridiales_Incertae_Sedis_XI | Clostridiales | Clostridia | Firmicutes |
| 8 | Fusobacterium | Fusobacteriaceae | Fusobacteriales | Fusobacteria | Fusobacteria |
| 9 | Propionibacterium | Propionibacteriaceae | Actinomycetales | Actinobacteria | Actinobacteria |
| 10 | Gemella | Bacillales_Incertae_Sedis_XI | Bacillales | Bacilli | Firmicutes |
| 11 | Rothia | Micrococcaceae | Actinomycetales | Actinobacteria | Actinobacteria |
| 12 | Granulicatella | Carnobacteriaceae | Lactobacillales | Bacilli | Firmicutes |
| 13 | Neisseria | Neisseriaceae | Neisseriales | Betaproteobacteria | Proteobacteria |
| 14 | Lactobacillus | Lactobacillaceae | Lactobacillales | Bacilli | Firmicutes |
| 15 | Megasphaera | Veillonellaceae | Selenomonadales | Negativicutes | Firmicutes |
| 16 | Catonella | Lachnospiraceae | Clostridiales | Clostridia | Firmicutes |
| 17 | Atopobium | Coriobacteriaceae | Coriobacteriales | Actinobacteria | Actinobacteria |
| 18 | Campylobacter | Campylobacteraceae | Campylobacterales | Epsilonproteobacteria | Proteobacteria |
| 19 | Capnocytophaga | Flavobacteriaceae | Flavobacteriales | Flavobacteria | Bacteroidetes |
| 20 | Solobacterium | Erysipelotrichaceae | Erysipelotrichales | Erysipelotrichia | Firmicutes |
| 21 | Moryella | Lachnospiraceae | Clostridiales | Clostridia | Firmicutes |
| 22 | TM7_genera_incertae_sedis | TM7_genera_incertae_sedis | TM7_genera_incertae_sedis | TM7_genera_incertae_sedis | TM7 |
| 23 | Staphylococcus | Staphylococcaceae | Bacillales | Bacilli | Firmicutes |
| 24 | Filifactor | Peptostreptococcaceae | Clostridiales | Clostridia | Firmicutes |
| 25 | Oribacterium | Lachnospiraceae | Clostridiales | Clostridia | Firmicutes |
| 26 | Burkholderia | Burkholderiaceae | Burkholderiales | Betaproteobacteria | Proteobacteria |
| 27 | Sneathia | Leptotrichiaceae | Fusobacteriales | Fusobacteria | Fusobacteria |
| 28 | Treponema | Spirochaetaceae | Spirochaetales | Spirochaetes | Spirochaetes |
| 29 | Moraxella | Moraxellaceae | Pseudomonadales | Gammaproteobacteria | Proteobacteria |
| 30 | Haemophilus | Pasteurellaceae | Pasteurellales | Gammaproteobacteria | Proteobacteria |
| 31 | Selenomonas | Veillonellaceae | Selenomonadales | Negativicutes | Firmicutes |
| 32 | Corynebacterium | Corynebacteriaceae | Actinomycetales | Actinobacteria | Actinobacteria |
| 33 | Rhizobium | Rhizobiaceae | Rhizobiales | Alphaproteobacteria | Proteobacteria |
| 34 | Bradyrhizobium | Bradyrhizobiaceae | Rhizobiales | Alphaproteobacteria | Proteobacteria |
| 35 | Methylobacterium | Methylobacteriaceae | Rhizobiales | Alphaproteobacteria | Proteobacteria |
| 36 | OD1_genera_incertae_sedis | OD1_genera_incertae_sedis | OD1_genera_incertae_sedis | OD1_genera_incertae_sedis | OD1 |
| 37 | Finegoldia | Clostridiales_Incertae_Sedis_XI | Clostridiales | Clostridia | Firmicutes |
| 38 | Microbacterium | Microbacteriaceae | Actinomycetales | Actinobacteria | Actinobacteria |
| 39 | Sphingomonas | Sphingomonadaceae | Sphingomonadales | Alphaproteobacteria | Proteobacteria |
| 40 | Chryseobacterium | Flavobacteriaceae | Flavobacteriales | Flavobacteria | Bacteroidetes |
| 41 | Bacteroides | Bacteroidaceae | Bacteroidales | Bacteroidia | Bacteroidetes |
| 42 | Bdellovibrio | Bdellovibrionaceae | Bdellovibrionales | Deltaproteobacteria | Proteobacteria |
| 43 | Streptophyta | Chloroplast | Chloroplast | Chloroplast | Cyanobacteria_Chloroplast |
| 44 | Lachnospiracea_incertae_sedis | Lachnospiraceae | Clostridiales | Clostridia | Firmicutes |
| 45 | Paracoccus | Rhodobacteraceae | Rhodobacterales | Alphaproteobacteria | Proteobacteria |
| 46 | Fastidiosipila | Ruminococcaceae | Clostridiales | Clostridia | Firmicutes |
| 47 | Pseudonocardia | Pseudonocardiaceae | Actinomycetales | Actinobacteria | Actinobacteria |
This file has .csv format below:
[3]:
with open(resource_filename('deepbiome', 'tests/data/genus48_dic.csv')) as f:
print(f.read())
Genus,Family,Order,Class,Phylum
Streptococcus,Streptococcaceae,Lactobacillales,Bacilli,Firmicutes
Tropheryma,Cellulomonadaceae,Actinomycetales,Actinobacteria,Actinobacteria
Veillonella,Veillonellaceae,Selenomonadales,Negativicutes,Firmicutes
Actinomyces,Actinomycetaceae,Actinomycetales,Actinobacteria,Actinobacteria
Flavobacterium,Flavobacteriaceae,Flavobacteriales,Flavobacteria,Bacteroidetes
Prevotella,Prevotellaceae,Bacteroidales,Bacteroidia,Bacteroidetes
Porphyromonas,Porphyromonadaceae,Bacteroidales,Bacteroidia,Bacteroidetes
Parvimonas,Clostridiales_Incertae_Sedis_XI,Clostridiales,Clostridia,Firmicutes
Fusobacterium,Fusobacteriaceae,Fusobacteriales,Fusobacteria,Fusobacteria
Propionibacterium,Propionibacteriaceae,Actinomycetales,Actinobacteria,Actinobacteria
Gemella,Bacillales_Incertae_Sedis_XI,Bacillales,Bacilli,Firmicutes
Rothia,Micrococcaceae,Actinomycetales,Actinobacteria,Actinobacteria
Granulicatella,Carnobacteriaceae,Lactobacillales,Bacilli,Firmicutes
Neisseria,Neisseriaceae,Neisseriales,Betaproteobacteria,Proteobacteria
Lactobacillus,Lactobacillaceae,Lactobacillales,Bacilli,Firmicutes
Megasphaera,Veillonellaceae,Selenomonadales,Negativicutes,Firmicutes
Catonella,Lachnospiraceae,Clostridiales,Clostridia,Firmicutes
Atopobium,Coriobacteriaceae,Coriobacteriales,Actinobacteria,Actinobacteria
Campylobacter,Campylobacteraceae,Campylobacterales,Epsilonproteobacteria,Proteobacteria
Capnocytophaga,Flavobacteriaceae,Flavobacteriales,Flavobacteria,Bacteroidetes
Solobacterium,Erysipelotrichaceae,Erysipelotrichales,Erysipelotrichia,Firmicutes
Moryella,Lachnospiraceae,Clostridiales,Clostridia,Firmicutes
TM7_genera_incertae_sedis,TM7_genera_incertae_sedis,TM7_genera_incertae_sedis,TM7_genera_incertae_sedis,TM7
Staphylococcus,Staphylococcaceae,Bacillales,Bacilli,Firmicutes
Filifactor,Peptostreptococcaceae,Clostridiales,Clostridia,Firmicutes
Oribacterium,Lachnospiraceae,Clostridiales,Clostridia,Firmicutes
Burkholderia,Burkholderiaceae,Burkholderiales,Betaproteobacteria,Proteobacteria
Sneathia,Leptotrichiaceae,Fusobacteriales,Fusobacteria,Fusobacteria
Treponema,Spirochaetaceae,Spirochaetales,Spirochaetes,Spirochaetes
Moraxella,Moraxellaceae,Pseudomonadales,Gammaproteobacteria,Proteobacteria
Haemophilus,Pasteurellaceae,Pasteurellales,Gammaproteobacteria,Proteobacteria
Selenomonas,Veillonellaceae,Selenomonadales,Negativicutes,Firmicutes
Corynebacterium,Corynebacteriaceae,Actinomycetales,Actinobacteria,Actinobacteria
Rhizobium,Rhizobiaceae,Rhizobiales,Alphaproteobacteria,Proteobacteria
Bradyrhizobium,Bradyrhizobiaceae,Rhizobiales,Alphaproteobacteria,Proteobacteria
Methylobacterium,Methylobacteriaceae,Rhizobiales,Alphaproteobacteria,Proteobacteria
OD1_genera_incertae_sedis,OD1_genera_incertae_sedis,OD1_genera_incertae_sedis,OD1_genera_incertae_sedis,OD1
Finegoldia,Clostridiales_Incertae_Sedis_XI,Clostridiales,Clostridia,Firmicutes
Microbacterium,Microbacteriaceae,Actinomycetales,Actinobacteria,Actinobacteria
Sphingomonas,Sphingomonadaceae,Sphingomonadales,Alphaproteobacteria,Proteobacteria
Chryseobacterium,Flavobacteriaceae,Flavobacteriales,Flavobacteria,Bacteroidetes
Bacteroides,Bacteroidaceae,Bacteroidales,Bacteroidia,Bacteroidetes
Bdellovibrio,Bdellovibrionaceae,Bdellovibrionales,Deltaproteobacteria,Proteobacteria
Streptophyta,Chloroplast,Chloroplast,Chloroplast,Cyanobacteria_Chloroplast
Lachnospiracea_incertae_sedis,Lachnospiraceae,Clostridiales,Clostridia,Firmicutes
Paracoccus,Rhodobacteraceae,Rhodobacterales,Alphaproteobacteria,Proteobacteria
Fastidiosipila,Ruminococcaceae,Clostridiales,Clostridia,Firmicutes
Pseudonocardia,Pseudonocardiaceae,Actinomycetales,Actinobacteria,Actinobacteria
Example of the list of the name of input files¶
In this example. we assume that input is given by the lists of files. Each file has all sample’s information for one repeatition. If we want to use the list of the input files, we need to make a list of the names of each input file. Below is an example file for k=3 repetition.
[4]:
list_of_input_files = pd.read_csv(resource_filename('deepbiome', 'tests/data/gcount_list.csv'))
list_of_input_files
[4]:
| 0 | |
|---|---|
| 0 | gcount_0001.csv |
| 1 | gcount_0002.csv |
| 2 | gcount_0003.csv |
Example of the lists of the input files¶
Below is an example of each input file. This example has 1000 samples as rows, and the abandunces of each microbiomes as columns. Below is an example file for k=3 repetition. This example is gcount_0001.csv for the first repetition in the list of the names of input files above. This file has the 1000 samples’ microbiome abandunce. The order of the microbiome should be same as the order of the microbiome in the Genus level in the tree information above.
[5]:
x_1 = pd.read_csv(resource_filename('deepbiome', 'tests/data/count/%s' % list_of_input_files.iloc[0,0]))
x_1.head()
[5]:
| Streptococcus | Tropheryma | Veillonella | Actinomyces | Flavobacterium | Prevotella | Porphyromonas | Parvimonas | Fusobacterium | Propionibacterium | ... | Microbacterium | Sphingomonas | Chryseobacterium | Bacteroides | Bdellovibrio | Streptophyta | Lachnospiracea_incertae_sedis | Paracoccus | Fastidiosipila | Pseudonocardia | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 841 | 0 | 813 | 505 | 5 | 3224 | 0 | 362 | 11 | 65 | ... | 0 | 87 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2133 |
| 1 | 1445 | 0 | 1 | 573 | 0 | 1278 | 82 | 85 | 69 | 154 | ... | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 3638 |
| 2 | 1259 | 0 | 805 | 650 | 0 | 1088 | 0 | 0 | 74 | 0 | ... | 0 | 2 | 8 | 1 | 39 | 0 | 0 | 0 | 0 | 3445 |
| 3 | 982 | 0 | 327 | 594 | 0 | 960 | 81 | 19 | 9 | 0 | ... | 157 | 1 | 0 | 4 | 60 | 0 | 0 | 0 | 0 | 3507 |
| 4 | 1162 | 0 | 130 | 969 | 163 | 1515 | 167 | 4 | 162 | 3 | ... | 0 | 9 | 0 | 0 | 0 | 0 | 60 | 0 | 0 | 3945 |
5 rows × 48 columns
[6]:
x_1.tail()
[6]:
| Streptococcus | Tropheryma | Veillonella | Actinomyces | Flavobacterium | Prevotella | Porphyromonas | Parvimonas | Fusobacterium | Propionibacterium | ... | Microbacterium | Sphingomonas | Chryseobacterium | Bacteroides | Bdellovibrio | Streptophyta | Lachnospiracea_incertae_sedis | Paracoccus | Fastidiosipila | Pseudonocardia | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 995 | 1401 | 4 | 30 | 526 | 0 | 923 | 25 | 0 | 127 | 0 | ... | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 4470 |
| 996 | 2655 | 6 | 106 | 74 | 0 | 952 | 76 | 13 | 158 | 125 | ... | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2826 |
| 997 | 335 | 0 | 71 | 259 | 67 | 718 | 1 | 4 | 4 | 167 | ... | 0 | 246 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 6527 |
| 998 | 649 | 69 | 966 | 1227 | 0 | 508 | 2 | 30 | 550 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 4402 |
| 999 | 1258 | 0 | 0 | 1119 | 0 | 2348 | 25 | 0 | 137 | 176 | ... | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2585 |
5 rows × 48 columns
This file has .csv format below:
[7]:
with open(resource_filename('deepbiome', 'tests/data/count/%s' % list_of_input_files.iloc[0,0])) as f:
x_csv = f.readlines()
_ = [print(l) for l in x_csv[:10]]
"Streptococcus","Tropheryma","Veillonella","Actinomyces","Flavobacterium","Prevotella","Porphyromonas","Parvimonas","Fusobacterium","Propionibacterium","Gemella","Rothia","Granulicatella","Neisseria","Lactobacillus","Megasphaera","Catonella","Atopobium","Campylobacter","Capnocytophaga","Solobacterium","Moryella","TM7_genera_incertae_sedis","Staphylococcus","Filifactor","Oribacterium","Burkholderia","Sneathia","Treponema","Moraxella","Haemophilus","Selenomonas","Corynebacterium","Rhizobium","Bradyrhizobium","Methylobacterium","OD1_genera_incertae_sedis","Finegoldia","Microbacterium","Sphingomonas","Chryseobacterium","Bacteroides","Bdellovibrio","Streptophyta","Lachnospiracea_incertae_sedis","Paracoccus","Fastidiosipila","Pseudonocardia"
841,0,813,505,5,3224,0,362,11,65,156,1,55,0,1,20,382,1,333,24,80,43,309,2,3,4,0,1,32,0,2,4,382,0,0,96,23,0,0,87,0,0,0,0,0,0,0,2133
1445,0,1,573,0,1278,82,85,69,154,436,3,0,61,440,0,394,83,33,123,0,49,414,0,0,37,0,0,42,0,0,384,27,0,0,0,146,0,0,1,2,0,0,0,0,0,0,3638
1259,0,805,650,0,1088,0,0,74,0,155,228,430,765,0,0,11,102,68,90,77,83,322,10,0,7,0,122,76,0,1,25,0,0,0,44,13,0,0,2,8,1,39,0,0,0,0,3445
982,0,327,594,0,960,81,19,9,0,45,457,1049,0,3,450,19,170,388,147,0,0,41,63,0,1,0,0,121,0,0,1,0,0,0,0,344,0,157,1,0,4,60,0,0,0,0,3507
1162,0,130,969,163,1515,167,4,162,3,12,0,48,73,93,259,52,0,201,85,14,14,434,2,0,0,0,0,187,0,0,188,45,0,0,0,4,0,0,9,0,0,0,0,60,0,0,3945
1956,37,41,661,47,1555,374,7,142,19,61,226,0,30,52,0,6,480,142,148,9,575,12,0,0,0,0,3,168,0,56,50,0,0,0,98,989,0,0,12,0,0,0,0,0,0,0,2044
1037,14,83,1595,132,305,103,174,1195,0,410,224,1,320,26,0,476,0,7,37,46,61,20,0,0,0,0,0,226,0,239,8,1,0,0,0,0,188,0,20,4,0,4,0,0,0,0,3044
641,0,172,179,0,1312,84,9,81,376,128,223,160,0,532,155,89,355,1,282,0,0,25,0,0,43,0,9,311,0,0,0,0,0,0,0,845,0,0,8,0,0,0,0,0,0,0,3980
852,146,504,99,2,376,116,152,67,0,120,3,23,2,34,0,127,75,240,60,42,0,9,0,15,0,62,0,13,0,197,187,396,0,0,20,51,0,0,3,0,0,0,0,0,0,0,6007
Example of the Y (regression)¶
This is an example of the output file for regression problem. One column contains outputs of the samples for one repetition. Below example file has 1000 samples in rows, k=3 repetitions in columns.
[8]:
y = pd.read_csv(resource_filename('deepbiome', 'tests/data/regression_y.csv'))
y.head()
[8]:
| x1 | x2 | x3 | |
|---|---|---|---|
| 0 | 4.997 | 5.492 | 5.474 |
| 1 | 5.004 | 1.500 | 4.640 |
| 2 | 5.485 | 4.187 | 5.491 |
| 3 | 5.490 | 4.863 | 1.500 |
| 4 | 1.500 | 5.481 | 5.490 |
[9]:
y.tail()
[9]:
| x1 | x2 | x3 | |
|---|---|---|---|
| 995 | 2.610 | 5.491 | 3.319 |
| 996 | 5.489 | 3.740 | 5.489 |
| 997 | 3.498 | 4.250 | 5.488 |
| 998 | 5.486 | 1.917 | 5.415 |
| 999 | 5.320 | 5.483 | 1.500 |
For one repetition, the DeepBiome will use the one column.
[10]:
y.iloc[:,0].head()
[10]:
0 4.997
1 5.004
2 5.485
3 5.490
4 1.500
Name: x1, dtype: float64
[11]:
y.iloc[:,0].tail()
[11]:
995 2.610
996 5.489
997 3.498
998 5.486
999 5.320
Name: x1, dtype: float64
Example of the Y (classification)¶
This is an example of the output file for classification problem. Below example file has 1000 samples in rows, 3 repetitions in columns.
[12]:
y = pd.read_csv(resource_filename('deepbiome', 'tests/data/classification_y.csv'))
y.head()
[12]:
| V1 | V2 | V3 | |
|---|---|---|---|
| 0 | 1 | 0 | 1 |
| 1 | 1 | 1 | 1 |
| 2 | 0 | 1 | 0 |
| 3 | 0 | 1 | 1 |
| 4 | 1 | 1 | 0 |
[13]:
y.tail()
[13]:
| V1 | V2 | V3 | |
|---|---|---|---|
| 995 | 1 | 0 | 1 |
| 996 | 0 | 1 | 0 |
| 997 | 1 | 1 | 0 |
| 998 | 0 | 1 | 1 |
| 999 | 1 | 1 | 1 |
For one repetition, the DeepBiome will use the one column.
[14]:
y.iloc[:,0].head()
[14]:
0 1
1 1
2 0
3 0
4 1
Name: V1, dtype: int64
[15]:
y.iloc[:,0].tail()
[15]:
995 1
996 0
997 1
998 0
999 1
Name: V1, dtype: int64
Exmple of the training index file for repetition¶
For each repetition, we have to set the training and test set. If the index file is given, DeepBiome sets the training set and test set based on the index file. Below is the example of the index file. Each column has the training indexs for each repetition. The DeepBiome will only use the samples in this index set for training.
[16]:
idxs = pd.read_csv(resource_filename('deepbiome', 'tests/data/regression_idx.csv'), dtype=np.int)
idxs.head()
[16]:
| V1 | V2 | V3 | |
|---|---|---|---|
| 0 | 490 | 690 | 62 |
| 1 | 498 | 968 | 123 |
| 2 | 389 | 999 | 335 |
| 3 | 51 | 139 | 843 |
| 4 | 592 | 83 | 204 |
[17]:
idxs.tail()
[17]:
| V1 | V2 | V3 | |
|---|---|---|---|
| 745 | 599 | 824 | 997 |
| 746 | 720 | 633 | 821 |
| 747 | 80 | 268 | 661 |
| 748 | 570 | 32 | 750 |
| 749 | 440 | 589 | 607 |
Below is the index set for 1st repetition. From 1000 samples above, it uses 750 samples for training.
[18]:
idxs.iloc[:,0].head()
[18]:
0 490
1 498
2 389
3 51
4 592
Name: V1, dtype: int64
[19]:
idxs.iloc[:,0].tail()
[19]:
745 599
746 720
747 80
748 570
749 440
Name: V1, dtype: int64
3. Prepare the configuration¶
For detailed configuration, we can build the configuration information for the network training by: 1. the python dictionary format 1. the configufation file (.cfg).
In this notebook, we show the python dictionary format configuration.
Please check the detailed information about each option in the documantation
For preparing the configuration about the network information (network_info)¶
To give the information about the training process, we provide a dictionary of configurations to the netowrk_info field. Your configuration for the network training should include the information about:
[20]:
network_info = {
'architecture_info': {
'batch_normalization': 'False',
'drop_out': '0',
'weight_initial': 'glorot_uniform',
'weight_l1_penalty':'0.',
'weight_decay': 'phylogenetic_tree',
},
'model_info': {
'lr': '0.01',
'decay': '0.001',
'loss': 'binary_crossentropy',
'metrics': 'binary_accuracy, sensitivity, specificity, gmeasure, auc',
'taxa_selection_metrics': 'sensitivity, specificity, gmeasure, accuracy',
'network_class': 'DeepBiomeNetwork',
'optimizer': 'adam',
'reader_class': 'MicroBiomeClassificationReader',
'normalizer': 'normalize_minmax',
},
'training_info': {
'batch_size': '50',
'epochs': '10',
'callbacks': 'ModelCheckpoint',
'monitor': 'val_loss',
'mode' : 'min',
'min_delta': '1e-7',
},
'validation_info': {
'batch_size': 'None',
'validation_size': '0.2'
},
'test_info': {
'batch_size': 'None'
}
}
For preparing the configuration about the path information (path_info)¶
To give the information about the path of dataset, paths for saving the trained weights and the evaluation results, we provide a dictionary of configurations to the path_info field. Your configuration for the network training should include the information about:
[21]:
path_info = {
'data_info': {
'count_list_path': resource_filename('deepbiome', 'tests/data/gcount_list.csv'),
'count_path': resource_filename('deepbiome', 'tests/data/count'),
'data_path': resource_filename('deepbiome', 'tests/data'),
'idx_path': resource_filename('deepbiome', 'tests/data/classification_idx.csv'),
'tree_info_path': resource_filename('deepbiome', 'tests/data/genus48_dic.csv'),
'x_path': '',
'y_path': 'classification_y.csv'
},
'model_info': {
'evaluation': 'eval.npy',
'history': 'hist.json',
'model_dir': './example_result/',
'weight': 'weight.h5'
}
}
4. DeepBiome Training¶
Now we can train the DeepBiome network based on the configurations.
For logging, we use the python logging library.
[22]:
logging.basicConfig(format = '[%(name)-8s|%(levelname)s|%(filename)s:%(lineno)s] %(message)s',
level=logging.DEBUG)
log = logging.getLogger()
The deeobiome_train function provide the test evaluation, train evaluation and the deepbiome network instance.
If we set number_of_fold, then DeepBiome performs cross-validation based on that value. If not, DeepBiome package performs cross-validation based on the index file. If both number_of_fold option and the index file are missing, then the library performs leave-one-out-cross-validation (LOOCV).
[23]:
test_evaluation, train_evaluation, network = deepbiome.deepbiome_train(log, network_info, path_info, number_of_fold=None)
[root |INFO|deepbiome.py:115] -----------------------------------------------------------------
[root |INFO|deepbiome.py:153] -------1 simulation start!----------------------------------
[root |INFO|readers.py:58] -----------------------------------------------------------------------
[root |INFO|readers.py:59] Construct Dataset
[root |INFO|readers.py:60] -----------------------------------------------------------------------
[root |INFO|readers.py:61] Load data
[root |INFO|deepbiome.py:164] -----------------------------------------------------------------
[root |INFO|deepbiome.py:165] Build network for 1 simulation
[root |INFO|build_network.py:521] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:522] Read phylogenetic tree information from /DATA/home/muha/github_repos/deepbiome/deepbiome/tests/data/genus48_dic.csv
[root |INFO|build_network.py:528] Phylogenetic tree level list: ['Genus', 'Family', 'Order', 'Class', 'Phylum']
[root |INFO|build_network.py:529] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:537] Genus: 48
[root |INFO|build_network.py:537] Family: 40
[root |INFO|build_network.py:537] Order: 23
[root |INFO|build_network.py:537] Class: 17
[root |INFO|build_network.py:537] Phylum: 9
[root |INFO|build_network.py:546] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:547] Phylogenetic_tree_dict info: ['Phylum', 'Order', 'Family', 'Class', 'Number', 'Genus']
[root |INFO|build_network.py:548] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:558] Build edge weights between [ Genus, Family]
[root |INFO|build_network.py:558] Build edge weights between [Family, Order]
[root |INFO|build_network.py:558] Build edge weights between [ Order, Class]
[root |INFO|build_network.py:558] Build edge weights between [ Class, Phylum]
[root |INFO|build_network.py:571] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:586] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:587] Build network based on phylogenetic tree information
[root |INFO|build_network.py:588] ------------------------------------------------------------------------------------------
WARNING:tensorflow:From /usr/local/lib/python3.5/dist-packages/tensorflow/python/ops/resource_variable_ops.py:432: colocate_with (from tensorflow.python.framework.ops) is deprecated and will be removed in a future version.
Instructions for updating:
Colocations handled automatically by placer.
[tensorflow|WARNING|deprecation.py:328] From /usr/local/lib/python3.5/dist-packages/tensorflow/python/ops/resource_variable_ops.py:432: colocate_with (from tensorflow.python.framework.ops) is deprecated and will be removed in a future version.
Instructions for updating:
Colocations handled automatically by placer.
[root |INFO|build_network.py:670] ------------------------------------------------------------------------------------------
Model: "model_1"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input (InputLayer) (None, 48) 0
_________________________________________________________________
l1_dense (Dense_with_tree) (None, 40) 1960
_________________________________________________________________
l1_activation (Activation) (None, 40) 0
_________________________________________________________________
l2_dense (Dense_with_tree) (None, 23) 943
_________________________________________________________________
l2_activation (Activation) (None, 23) 0
_________________________________________________________________
l3_dense (Dense_with_tree) (None, 17) 408
_________________________________________________________________
l3_activation (Activation) (None, 17) 0
_________________________________________________________________
l4_dense (Dense_with_tree) (None, 9) 162
_________________________________________________________________
l4_activation (Activation) (None, 9) 0
_________________________________________________________________
last_dense_h (Dense) (None, 1) 10
_________________________________________________________________
p_hat (Activation) (None, 1) 0
=================================================================
Total params: 3,483
Trainable params: 3,483
Non-trainable params: 0
_________________________________________________________________
[root |INFO|build_network.py:61] Build Network
[root |INFO|build_network.py:62] Optimizer = adam
[root |INFO|build_network.py:63] Loss = binary_crossentropy
[root |INFO|build_network.py:64] Metrics = binary_accuracy, sensitivity, specificity, gmeasure, auc
[root |INFO|deepbiome.py:176] -----------------------------------------------------------------
[root |INFO|deepbiome.py:177] 1 fold computing start!----------------------------------
[root |INFO|build_network.py:137] Training start!
WARNING:tensorflow:From /usr/local/lib/python3.5/dist-packages/tensorflow/python/ops/math_ops.py:2862: to_int32 (from tensorflow.python.ops.math_ops) is deprecated and will be removed in a future version.
Instructions for updating:
Use tf.cast instead.
[tensorflow|WARNING|deprecation.py:328] From /usr/local/lib/python3.5/dist-packages/tensorflow/python/ops/math_ops.py:2862: to_int32 (from tensorflow.python.ops.math_ops) is deprecated and will be removed in a future version.
Instructions for updating:
Use tf.cast instead.
Train on 600 samples, validate on 150 samples
Epoch 1/10
600/600 [==============================] - 1s 1ms/step - loss: 0.6743 - binary_accuracy: 0.6517 - sensitivity: 0.9212 - specificity: 0.0833 - gmeasure: 0.0194 - auc: 0.4725 - val_loss: 0.6409 - val_binary_accuracy: 0.7000 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.5159
Epoch 2/10
600/600 [==============================] - 0s 223us/step - loss: 0.6310 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - auc: 0.7404 - val_loss: 0.6117 - val_binary_accuracy: 0.7000 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.7595
Epoch 3/10
600/600 [==============================] - 0s 199us/step - loss: 0.6206 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - auc: 0.8101 - val_loss: 0.6110 - val_binary_accuracy: 0.7000 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.7792
Epoch 4/10
600/600 [==============================] - 0s 225us/step - loss: 0.6214 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - auc: 0.8168 - val_loss: 0.6108 - val_binary_accuracy: 0.7000 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.7948
Epoch 5/10
600/600 [==============================] - 0s 217us/step - loss: 0.6204 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - auc: 0.8246 - val_loss: 0.6112 - val_binary_accuracy: 0.7000 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.8087
Epoch 6/10
600/600 [==============================] - 0s 237us/step - loss: 0.6205 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - auc: 0.8379 - val_loss: 0.6113 - val_binary_accuracy: 0.7000 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.8203
Epoch 7/10
600/600 [==============================] - 0s 238us/step - loss: 0.6220 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - auc: 0.8410 - val_loss: 0.6123 - val_binary_accuracy: 0.7000 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.8255
Epoch 8/10
600/600 [==============================] - 0s 205us/step - loss: 0.6196 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - auc: 0.8492 - val_loss: 0.6103 - val_binary_accuracy: 0.7000 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.8313
Epoch 9/10
600/600 [==============================] - 0s 219us/step - loss: 0.6202 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - auc: 0.8526 - val_loss: 0.6098 - val_binary_accuracy: 0.7000 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.8407
Epoch 10/10
600/600 [==============================] - 0s 195us/step - loss: 0.6204 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - auc: 0.8572 - val_loss: 0.6092 - val_binary_accuracy: 0.7000 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.8457
[root |INFO|build_network.py:87] Load trained model weight at ./example_result/weight_0.h5
[root |INFO|build_network.py:147] Training end with time 4.45297384262085!
[root |INFO|build_network.py:83] Saved trained model weight at ./example_result/weight_0.h5
[root |DEBUG|deepbiome.py:185] Save weight at ./example_result/weight_0.h5
[root |DEBUG|deepbiome.py:188] Save history at ./example_result/hist_0.json
[root |INFO|build_network.py:173] Evaluation start!
750/750 [==============================] - 0s 7us/step
[root |INFO|build_network.py:178] Evaluation end with time 0.011869668960571289!
[root |INFO|build_network.py:179] Evaluation: [0.6164737343788147, 0.690666675567627, 1.0, 0.0, 0.0, 0.8536480069160461]
[root |INFO|build_network.py:173] Evaluation start!
250/250 [==============================] - 0s 18us/step
[root |INFO|build_network.py:178] Evaluation end with time 0.01761341094970703!
[root |INFO|build_network.py:179] Evaluation: [0.6278528571128845, 0.6759999990463257, 1.0, 0.0, 0.0, 0.8606910705566406]
[root |INFO|deepbiome.py:199] Compute time : 5.686180830001831
[root |INFO|deepbiome.py:200] 1 fold computing end!---------------------------------------------
[root |INFO|deepbiome.py:153] -------2 simulation start!----------------------------------
[root |INFO|readers.py:58] -----------------------------------------------------------------------
[root |INFO|readers.py:59] Construct Dataset
[root |INFO|readers.py:60] -----------------------------------------------------------------------
[root |INFO|readers.py:61] Load data
[root |INFO|deepbiome.py:164] -----------------------------------------------------------------
[root |INFO|deepbiome.py:165] Build network for 2 simulation
[root |INFO|build_network.py:521] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:522] Read phylogenetic tree information from /DATA/home/muha/github_repos/deepbiome/deepbiome/tests/data/genus48_dic.csv
[root |INFO|build_network.py:528] Phylogenetic tree level list: ['Genus', 'Family', 'Order', 'Class', 'Phylum']
[root |INFO|build_network.py:529] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:537] Genus: 48
[root |INFO|build_network.py:537] Family: 40
[root |INFO|build_network.py:537] Order: 23
[root |INFO|build_network.py:537] Class: 17
[root |INFO|build_network.py:537] Phylum: 9
[root |INFO|build_network.py:546] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:547] Phylogenetic_tree_dict info: ['Phylum', 'Order', 'Family', 'Class', 'Number', 'Genus']
[root |INFO|build_network.py:548] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:558] Build edge weights between [ Genus, Family]
[root |INFO|build_network.py:558] Build edge weights between [Family, Order]
[root |INFO|build_network.py:558] Build edge weights between [ Order, Class]
[root |INFO|build_network.py:558] Build edge weights between [ Class, Phylum]
[root |INFO|build_network.py:571] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:586] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:587] Build network based on phylogenetic tree information
[root |INFO|build_network.py:588] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:670] ------------------------------------------------------------------------------------------
Model: "model_1"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input (InputLayer) (None, 48) 0
_________________________________________________________________
l1_dense (Dense_with_tree) (None, 40) 1960
_________________________________________________________________
l1_activation (Activation) (None, 40) 0
_________________________________________________________________
l2_dense (Dense_with_tree) (None, 23) 943
_________________________________________________________________
l2_activation (Activation) (None, 23) 0
_________________________________________________________________
l3_dense (Dense_with_tree) (None, 17) 408
_________________________________________________________________
l3_activation (Activation) (None, 17) 0
_________________________________________________________________
l4_dense (Dense_with_tree) (None, 9) 162
_________________________________________________________________
l4_activation (Activation) (None, 9) 0
_________________________________________________________________
last_dense_h (Dense) (None, 1) 10
_________________________________________________________________
p_hat (Activation) (None, 1) 0
=================================================================
Total params: 3,483
Trainable params: 3,483
Non-trainable params: 0
_________________________________________________________________
[root |INFO|build_network.py:61] Build Network
[root |INFO|build_network.py:62] Optimizer = adam
[root |INFO|build_network.py:63] Loss = binary_crossentropy
[root |INFO|build_network.py:64] Metrics = binary_accuracy, sensitivity, specificity, gmeasure, auc
[root |INFO|deepbiome.py:176] -----------------------------------------------------------------
[root |INFO|deepbiome.py:177] 2 fold computing start!----------------------------------
[root |INFO|build_network.py:137] Training start!
Train on 600 samples, validate on 150 samples
Epoch 1/10
600/600 [==============================] - 1s 1ms/step - loss: 0.6452 - binary_accuracy: 0.7067 - sensitivity: 0.9363 - specificity: 0.0677 - gmeasure: 0.0364 - auc: 0.4412 - val_loss: 0.6043 - val_binary_accuracy: 0.7267 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.5880
Epoch 2/10
600/600 [==============================] - 0s 613us/step - loss: 0.5917 - binary_accuracy: 0.7283 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - auc: 0.5369 - val_loss: 0.5893 - val_binary_accuracy: 0.7267 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.6307
Epoch 3/10
600/600 [==============================] - 0s 289us/step - loss: 0.5877 - binary_accuracy: 0.7283 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - auc: 0.6219 - val_loss: 0.5875 - val_binary_accuracy: 0.7267 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.6821
Epoch 4/10
600/600 [==============================] - 0s 621us/step - loss: 0.5869 - binary_accuracy: 0.7283 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - auc: 0.6838 - val_loss: 0.5867 - val_binary_accuracy: 0.7267 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.7095
Epoch 5/10
600/600 [==============================] - 0s 335us/step - loss: 0.5855 - binary_accuracy: 0.7283 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - auc: 0.7137 - val_loss: 0.5867 - val_binary_accuracy: 0.7267 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.7346
Epoch 6/10
600/600 [==============================] - 0s 597us/step - loss: 0.5860 - binary_accuracy: 0.7283 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - auc: 0.7500 - val_loss: 0.5866 - val_binary_accuracy: 0.7267 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.7710
Epoch 7/10
600/600 [==============================] - 0s 223us/step - loss: 0.5852 - binary_accuracy: 0.7283 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - auc: 0.7697 - val_loss: 0.5867 - val_binary_accuracy: 0.7267 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.7906
Epoch 8/10
600/600 [==============================] - 0s 452us/step - loss: 0.5855 - binary_accuracy: 0.7283 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - auc: 0.8008 - val_loss: 0.5869 - val_binary_accuracy: 0.7267 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.8195
Epoch 9/10
600/600 [==============================] - 0s 471us/step - loss: 0.5878 - binary_accuracy: 0.7283 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - auc: 0.8200 - val_loss: 0.5871 - val_binary_accuracy: 0.7267 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.8315
Epoch 10/10
600/600 [==============================] - 0s 390us/step - loss: 0.5874 - binary_accuracy: 0.7283 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - auc: 0.8272 - val_loss: 0.5868 - val_binary_accuracy: 0.7267 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.8467
[root |INFO|build_network.py:87] Load trained model weight at ./example_result/weight_1.h5
[root |INFO|build_network.py:147] Training end with time 4.857975244522095!
[root |INFO|build_network.py:83] Saved trained model weight at ./example_result/weight_1.h5
[root |DEBUG|deepbiome.py:185] Save weight at ./example_result/weight_1.h5
[root |DEBUG|deepbiome.py:188] Save history at ./example_result/hist_1.json
[root |INFO|build_network.py:173] Evaluation start!
750/750 [==============================] - 0s 18us/step
[root |INFO|build_network.py:178] Evaluation end with time 0.028972148895263672!
[root |INFO|build_network.py:179] Evaluation: [0.5853651762008667, 0.7279999852180481, 1.0, 0.0, 0.0, 0.7630988359451294]
[root |INFO|build_network.py:173] Evaluation start!
250/250 [==============================] - 0s 48us/step
[root |INFO|build_network.py:178] Evaluation end with time 0.02693629264831543!
[root |INFO|build_network.py:179] Evaluation: [0.6156458258628845, 0.6959999799728394, 1.0, 0.0, 0.0, 0.782667875289917]
[root |INFO|deepbiome.py:199] Compute time : 5.397497892379761
[root |INFO|deepbiome.py:200] 2 fold computing end!---------------------------------------------
[root |INFO|deepbiome.py:153] -------3 simulation start!----------------------------------
[root |INFO|readers.py:58] -----------------------------------------------------------------------
[root |INFO|readers.py:59] Construct Dataset
[root |INFO|readers.py:60] -----------------------------------------------------------------------
[root |INFO|readers.py:61] Load data
[root |INFO|deepbiome.py:164] -----------------------------------------------------------------
[root |INFO|deepbiome.py:165] Build network for 3 simulation
[root |INFO|build_network.py:521] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:522] Read phylogenetic tree information from /DATA/home/muha/github_repos/deepbiome/deepbiome/tests/data/genus48_dic.csv
[root |INFO|build_network.py:528] Phylogenetic tree level list: ['Genus', 'Family', 'Order', 'Class', 'Phylum']
[root |INFO|build_network.py:529] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:537] Genus: 48
[root |INFO|build_network.py:537] Family: 40
[root |INFO|build_network.py:537] Order: 23
[root |INFO|build_network.py:537] Class: 17
[root |INFO|build_network.py:537] Phylum: 9
[root |INFO|build_network.py:546] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:547] Phylogenetic_tree_dict info: ['Phylum', 'Order', 'Family', 'Class', 'Number', 'Genus']
[root |INFO|build_network.py:548] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:558] Build edge weights between [ Genus, Family]
[root |INFO|build_network.py:558] Build edge weights between [Family, Order]
[root |INFO|build_network.py:558] Build edge weights between [ Order, Class]
[root |INFO|build_network.py:558] Build edge weights between [ Class, Phylum]
[root |INFO|build_network.py:571] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:586] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:587] Build network based on phylogenetic tree information
[root |INFO|build_network.py:588] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:670] ------------------------------------------------------------------------------------------
Model: "model_1"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input (InputLayer) (None, 48) 0
_________________________________________________________________
l1_dense (Dense_with_tree) (None, 40) 1960
_________________________________________________________________
l1_activation (Activation) (None, 40) 0
_________________________________________________________________
l2_dense (Dense_with_tree) (None, 23) 943
_________________________________________________________________
l2_activation (Activation) (None, 23) 0
_________________________________________________________________
l3_dense (Dense_with_tree) (None, 17) 408
_________________________________________________________________
l3_activation (Activation) (None, 17) 0
_________________________________________________________________
l4_dense (Dense_with_tree) (None, 9) 162
_________________________________________________________________
l4_activation (Activation) (None, 9) 0
_________________________________________________________________
last_dense_h (Dense) (None, 1) 10
_________________________________________________________________
p_hat (Activation) (None, 1) 0
=================================================================
Total params: 3,483
Trainable params: 3,483
Non-trainable params: 0
_________________________________________________________________
[root |INFO|build_network.py:61] Build Network
[root |INFO|build_network.py:62] Optimizer = adam
[root |INFO|build_network.py:63] Loss = binary_crossentropy
[root |INFO|build_network.py:64] Metrics = binary_accuracy, sensitivity, specificity, gmeasure, auc
[root |INFO|deepbiome.py:176] -----------------------------------------------------------------
[root |INFO|deepbiome.py:177] 3 fold computing start!----------------------------------
[root |INFO|build_network.py:137] Training start!
Train on 600 samples, validate on 150 samples
Epoch 1/10
600/600 [==============================] - 1s 1ms/step - loss: 0.6592 - binary_accuracy: 0.6867 - sensitivity: 0.9821 - specificity: 0.0189 - gmeasure: 0.0352 - auc: 0.5518 - val_loss: 0.6501 - val_binary_accuracy: 0.6533 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.5313
Epoch 2/10
600/600 [==============================] - 0s 203us/step - loss: 0.6245 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - auc: 0.5300 - val_loss: 0.6511 - val_binary_accuracy: 0.6533 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.5359
Epoch 3/10
600/600 [==============================] - 0s 186us/step - loss: 0.6232 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - auc: 0.5332 - val_loss: 0.6529 - val_binary_accuracy: 0.6533 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.5422
Epoch 4/10
600/600 [==============================] - 0s 211us/step - loss: 0.6226 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - auc: 0.5512 - val_loss: 0.6460 - val_binary_accuracy: 0.6533 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.5510
Epoch 5/10
600/600 [==============================] - 0s 183us/step - loss: 0.6222 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - auc: 0.5571 - val_loss: 0.6485 - val_binary_accuracy: 0.6533 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.5556
Epoch 6/10
600/600 [==============================] - 0s 179us/step - loss: 0.6216 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - auc: 0.5725 - val_loss: 0.6475 - val_binary_accuracy: 0.6533 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.5648
Epoch 7/10
600/600 [==============================] - 0s 224us/step - loss: 0.6206 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - auc: 0.5685 - val_loss: 0.6468 - val_binary_accuracy: 0.6533 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.5722
Epoch 8/10
600/600 [==============================] - 0s 214us/step - loss: 0.6201 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - auc: 0.5792 - val_loss: 0.6481 - val_binary_accuracy: 0.6533 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.5773
Epoch 9/10
600/600 [==============================] - 0s 239us/step - loss: 0.6203 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - auc: 0.5836 - val_loss: 0.6475 - val_binary_accuracy: 0.6533 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.5862
Epoch 10/10
600/600 [==============================] - 0s 171us/step - loss: 0.6205 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - auc: 0.5945 - val_loss: 0.6487 - val_binary_accuracy: 0.6533 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00 - val_auc: 0.5951
[root |INFO|build_network.py:87] Load trained model weight at ./example_result/weight_2.h5
[root |INFO|build_network.py:147] Training end with time 3.4070072174072266!
[root |INFO|build_network.py:83] Saved trained model weight at ./example_result/weight_2.h5
[root |DEBUG|deepbiome.py:185] Save weight at ./example_result/weight_2.h5
[root |DEBUG|deepbiome.py:188] Save history at ./example_result/hist_2.json
[root |INFO|build_network.py:173] Evaluation start!
750/750 [==============================] - 0s 9us/step
[root |INFO|build_network.py:178] Evaluation end with time 0.016149044036865234!
[root |INFO|build_network.py:179] Evaluation: [0.6257709264755249, 0.6813333630561829, 1.0, 0.0, 0.0, 0.5530750155448914]
[root |INFO|build_network.py:173] Evaluation start!
250/250 [==============================] - 0s 24us/step
[root |INFO|build_network.py:178] Evaluation end with time 0.014859676361083984!
[root |INFO|build_network.py:179] Evaluation: [0.6038214564323425, 0.7120000123977661, 1.0, 0.0, 0.0, 0.47752809524536133]
[root |INFO|deepbiome.py:199] Compute time : 4.102193593978882
[root |INFO|deepbiome.py:200] 3 fold computing end!---------------------------------------------
[root |INFO|deepbiome.py:211] -----------------------------------------------------------------
[root |INFO|deepbiome.py:212] Train Evaluation : ['loss' 'binary_accuracy' 'sensitivity' 'specificity' 'gmeasure' 'auc']
[root |INFO|deepbiome.py:213] mean : [0.609 0.700 1.000 0.000 0.000 0.723]
[root |INFO|deepbiome.py:214] std : [0.017 0.020 0.000 0.000 0.000 0.126]
[root |INFO|deepbiome.py:215] -----------------------------------------------------------------
[root |INFO|deepbiome.py:216] Test Evaluation : ['loss' 'binary_accuracy' 'sensitivity' 'specificity' 'gmeasure' 'auc']
[root |INFO|deepbiome.py:217] mean : [0.616 0.695 1.000 0.000 0.000 0.707]
[root |INFO|deepbiome.py:218] std : [0.010 0.015 0.000 0.000 0.000 0.165]
[root |INFO|deepbiome.py:219] -----------------------------------------------------------------
[root |INFO|deepbiome.py:230] Total Computing Ended
[root |INFO|deepbiome.py:231] -----------------------------------------------------------------
The deepbiome_train saves the trained model weights, evaluation results and history based on the path information from the configuration.
Lets check the history files.
[24]:
with open('./%s/hist_0.json' % path_info['model_info']['model_dir'], 'r') as f:
history = json.load(f)
plt.plot(history['val_loss'], label='Validation')
plt.plot(history['loss'], label='Training')
plt.legend()
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.show()
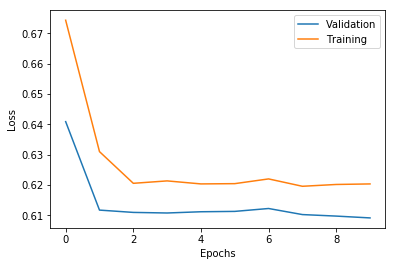
Test evauation and train evauation is the numpy array of the shape (number of repetitions, number of evaluation measures).
[25]:
test_evaluation
[25]:
array([[0.628, 0.676, 1.000, 0.000, 0.000, 0.861],
[0.616, 0.696, 1.000, 0.000, 0.000, 0.783],
[0.604, 0.712, 1.000, 0.000, 0.000, 0.478]])
[26]:
train_evaluation
[26]:
array([[0.616, 0.691, 1.000, 0.000, 0.000, 0.854],
[0.585, 0.728, 1.000, 0.000, 0.000, 0.763],
[0.626, 0.681, 1.000, 0.000, 0.000, 0.553]])
5. Load the pre-trained network for training¶
If you have a pre-trianed model, you warm_start next training using the pre-trained weights by setting the warm_start option in training_info to True. The file path of the pre-trained weights passed in the warm_start_model option. Below is the example:
[27]:
warm_start_network_info = {
'architecture_info': {
'batch_normalization': 'False',
'drop_out': '0',
'weight_initial': 'glorot_uniform',
'weight_l1_penalty':'0.',
'weight_decay': 'phylogenetic_tree',
},
'model_info': {
'decay': '0.001',
'loss': 'binary_crossentropy',
'lr': '0.01',
'metrics': 'binary_accuracy, sensitivity, specificity, gmeasure',
'network_class': 'DeepBiomeNetwork',
'normalizer': 'normalize_minmax',
'optimizer': 'adam',
'reader_class': 'MicroBiomeClassificationReader',
'taxa_selection_metrics': 'sensitivity, specificity, gmeasure, accuracy',
},
'training_info': {
'warm_start':'True',
'warm_start_model':'./example_result/weight.h5',
'batch_size': '50',
'epochs': '10',
'callbacks': 'ModelCheckpoint',
'monitor': 'val_loss',
'mode' : 'min',
'min_delta': '1e-7',
},
'validation_info': {
'batch_size': 'None',
'validation_size': '0.2'
},
'test_info': {
'batch_size': 'None'
}
}
[28]:
test_evaluation, train_evaluation, network = deepbiome.deepbiome_train(log, warm_start_network_info, path_info,
number_of_fold=None)
[root |INFO|deepbiome.py:115] -----------------------------------------------------------------
[root |INFO|deepbiome.py:153] -------1 simulation start!----------------------------------
[root |INFO|readers.py:58] -----------------------------------------------------------------------
[root |INFO|readers.py:59] Construct Dataset
[root |INFO|readers.py:60] -----------------------------------------------------------------------
[root |INFO|readers.py:61] Load data
[root |INFO|deepbiome.py:164] -----------------------------------------------------------------
[root |INFO|deepbiome.py:165] Build network for 1 simulation
[root |INFO|build_network.py:521] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:522] Read phylogenetic tree information from /DATA/home/muha/github_repos/deepbiome/deepbiome/tests/data/genus48_dic.csv
[root |INFO|build_network.py:528] Phylogenetic tree level list: ['Genus', 'Family', 'Order', 'Class', 'Phylum']
[root |INFO|build_network.py:529] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:537] Genus: 48
[root |INFO|build_network.py:537] Family: 40
[root |INFO|build_network.py:537] Order: 23
[root |INFO|build_network.py:537] Class: 17
[root |INFO|build_network.py:537] Phylum: 9
[root |INFO|build_network.py:546] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:547] Phylogenetic_tree_dict info: ['Phylum', 'Order', 'Family', 'Class', 'Number', 'Genus']
[root |INFO|build_network.py:548] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:558] Build edge weights between [ Genus, Family]
[root |INFO|build_network.py:558] Build edge weights between [Family, Order]
[root |INFO|build_network.py:558] Build edge weights between [ Order, Class]
[root |INFO|build_network.py:558] Build edge weights between [ Class, Phylum]
[root |INFO|build_network.py:571] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:586] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:587] Build network based on phylogenetic tree information
[root |INFO|build_network.py:588] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:670] ------------------------------------------------------------------------------------------
Model: "model_1"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input (InputLayer) (None, 48) 0
_________________________________________________________________
l1_dense (Dense_with_tree) (None, 40) 1960
_________________________________________________________________
l1_activation (Activation) (None, 40) 0
_________________________________________________________________
l2_dense (Dense_with_tree) (None, 23) 943
_________________________________________________________________
l2_activation (Activation) (None, 23) 0
_________________________________________________________________
l3_dense (Dense_with_tree) (None, 17) 408
_________________________________________________________________
l3_activation (Activation) (None, 17) 0
_________________________________________________________________
l4_dense (Dense_with_tree) (None, 9) 162
_________________________________________________________________
l4_activation (Activation) (None, 9) 0
_________________________________________________________________
last_dense_h (Dense) (None, 1) 10
_________________________________________________________________
p_hat (Activation) (None, 1) 0
=================================================================
Total params: 3,483
Trainable params: 3,483
Non-trainable params: 0
_________________________________________________________________
[root |INFO|build_network.py:61] Build Network
[root |INFO|build_network.py:62] Optimizer = adam
[root |INFO|build_network.py:63] Loss = binary_crossentropy
[root |INFO|build_network.py:64] Metrics = binary_accuracy, sensitivity, specificity, gmeasure
[root |INFO|build_network.py:87] Load trained model weight at ./example_result/weight_0.h5
[root |INFO|deepbiome.py:176] -----------------------------------------------------------------
[root |INFO|deepbiome.py:177] 1 fold computing start!----------------------------------
[root |INFO|build_network.py:137] Training start!
Train on 600 samples, validate on 150 samples
Epoch 1/10
600/600 [==============================] - 0s 797us/step - loss: 0.6207 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - val_loss: 0.6080 - val_binary_accuracy: 0.7000 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00
Epoch 2/10
600/600 [==============================] - 0s 123us/step - loss: 0.6181 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - val_loss: 0.6089 - val_binary_accuracy: 0.7000 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00
Epoch 3/10
600/600 [==============================] - 0s 107us/step - loss: 0.6151 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - val_loss: 0.6033 - val_binary_accuracy: 0.7000 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00
Epoch 4/10
600/600 [==============================] - 0s 118us/step - loss: 0.6123 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - val_loss: 0.5989 - val_binary_accuracy: 0.7000 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00
Epoch 5/10
600/600 [==============================] - 0s 126us/step - loss: 0.6060 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - val_loss: 0.5929 - val_binary_accuracy: 0.7000 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00
Epoch 6/10
600/600 [==============================] - 0s 149us/step - loss: 0.5992 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - val_loss: 0.5807 - val_binary_accuracy: 0.7000 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00
Epoch 7/10
600/600 [==============================] - 0s 144us/step - loss: 0.5838 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - val_loss: 0.5654 - val_binary_accuracy: 0.7000 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00
Epoch 8/10
600/600 [==============================] - 0s 165us/step - loss: 0.5646 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - val_loss: 0.5402 - val_binary_accuracy: 0.7000 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00
Epoch 9/10
600/600 [==============================] - 0s 158us/step - loss: 0.5396 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - val_loss: 0.5113 - val_binary_accuracy: 0.7000 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00
Epoch 10/10
600/600 [==============================] - 0s 136us/step - loss: 0.5159 - binary_accuracy: 0.7067 - sensitivity: 0.9975 - specificity: 0.0588 - gmeasure: 0.0983 - val_loss: 0.4866 - val_binary_accuracy: 0.7467 - val_sensitivity: 0.9447 - val_specificity: 0.2790 - val_gmeasure: 0.5111
[root |INFO|build_network.py:87] Load trained model weight at ./example_result/weight_0.h5
[root |INFO|build_network.py:147] Training end with time 2.830467700958252!
[root |INFO|build_network.py:83] Saved trained model weight at ./example_result/weight_0.h5
[root |DEBUG|deepbiome.py:185] Save weight at ./example_result/weight_0.h5
[root |DEBUG|deepbiome.py:188] Save history at ./example_result/hist_0.json
[root |INFO|build_network.py:173] Evaluation start!
750/750 [==============================] - 0s 3us/step
[root |INFO|build_network.py:178] Evaluation end with time 0.009613752365112305!
[root |INFO|build_network.py:179] Evaluation: [0.4993871748447418, 0.7493333220481873, 0.9517374634742737, 0.29741379618644714, 0.5320336818695068]
[root |INFO|build_network.py:173] Evaluation start!
250/250 [==============================] - 0s 8us/step
[root |INFO|build_network.py:178] Evaluation end with time 0.008378982543945312!
[root |INFO|build_network.py:179] Evaluation: [0.5000216364860535, 0.7480000257492065, 0.9585798978805542, 0.3086419701576233, 0.543928325176239]
[root |INFO|deepbiome.py:199] Compute time : 3.409234046936035
[root |INFO|deepbiome.py:200] 1 fold computing end!---------------------------------------------
[root |INFO|deepbiome.py:153] -------2 simulation start!----------------------------------
[root |INFO|readers.py:58] -----------------------------------------------------------------------
[root |INFO|readers.py:59] Construct Dataset
[root |INFO|readers.py:60] -----------------------------------------------------------------------
[root |INFO|readers.py:61] Load data
[root |INFO|deepbiome.py:164] -----------------------------------------------------------------
[root |INFO|deepbiome.py:165] Build network for 2 simulation
[root |INFO|build_network.py:521] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:522] Read phylogenetic tree information from /DATA/home/muha/github_repos/deepbiome/deepbiome/tests/data/genus48_dic.csv
[root |INFO|build_network.py:528] Phylogenetic tree level list: ['Genus', 'Family', 'Order', 'Class', 'Phylum']
[root |INFO|build_network.py:529] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:537] Genus: 48
[root |INFO|build_network.py:537] Family: 40
[root |INFO|build_network.py:537] Order: 23
[root |INFO|build_network.py:537] Class: 17
[root |INFO|build_network.py:537] Phylum: 9
[root |INFO|build_network.py:546] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:547] Phylogenetic_tree_dict info: ['Phylum', 'Order', 'Family', 'Class', 'Number', 'Genus']
[root |INFO|build_network.py:548] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:558] Build edge weights between [ Genus, Family]
[root |INFO|build_network.py:558] Build edge weights between [Family, Order]
[root |INFO|build_network.py:558] Build edge weights between [ Order, Class]
[root |INFO|build_network.py:558] Build edge weights between [ Class, Phylum]
[root |INFO|build_network.py:571] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:586] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:587] Build network based on phylogenetic tree information
[root |INFO|build_network.py:588] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:670] ------------------------------------------------------------------------------------------
Model: "model_1"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input (InputLayer) (None, 48) 0
_________________________________________________________________
l1_dense (Dense_with_tree) (None, 40) 1960
_________________________________________________________________
l1_activation (Activation) (None, 40) 0
_________________________________________________________________
l2_dense (Dense_with_tree) (None, 23) 943
_________________________________________________________________
l2_activation (Activation) (None, 23) 0
_________________________________________________________________
l3_dense (Dense_with_tree) (None, 17) 408
_________________________________________________________________
l3_activation (Activation) (None, 17) 0
_________________________________________________________________
l4_dense (Dense_with_tree) (None, 9) 162
_________________________________________________________________
l4_activation (Activation) (None, 9) 0
_________________________________________________________________
last_dense_h (Dense) (None, 1) 10
_________________________________________________________________
p_hat (Activation) (None, 1) 0
=================================================================
Total params: 3,483
Trainable params: 3,483
Non-trainable params: 0
_________________________________________________________________
[root |INFO|build_network.py:61] Build Network
[root |INFO|build_network.py:62] Optimizer = adam
[root |INFO|build_network.py:63] Loss = binary_crossentropy
[root |INFO|build_network.py:64] Metrics = binary_accuracy, sensitivity, specificity, gmeasure
[root |INFO|build_network.py:87] Load trained model weight at ./example_result/weight_1.h5
[root |INFO|deepbiome.py:176] -----------------------------------------------------------------
[root |INFO|deepbiome.py:177] 2 fold computing start!----------------------------------
[root |INFO|build_network.py:137] Training start!
Train on 600 samples, validate on 150 samples
Epoch 1/10
600/600 [==============================] - 0s 830us/step - loss: 0.5858 - binary_accuracy: 0.7283 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - val_loss: 0.5866 - val_binary_accuracy: 0.7267 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00
Epoch 2/10
600/600 [==============================] - 0s 159us/step - loss: 0.5853 - binary_accuracy: 0.7283 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - val_loss: 0.5865 - val_binary_accuracy: 0.7267 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00
Epoch 3/10
600/600 [==============================] - 0s 148us/step - loss: 0.5857 - binary_accuracy: 0.7283 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - val_loss: 0.5867 - val_binary_accuracy: 0.7267 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00
Epoch 4/10
600/600 [==============================] - 0s 126us/step - loss: 0.5862 - binary_accuracy: 0.7283 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - val_loss: 0.5869 - val_binary_accuracy: 0.7267 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00
Epoch 5/10
600/600 [==============================] - 0s 124us/step - loss: 0.5852 - binary_accuracy: 0.7283 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - val_loss: 0.5865 - val_binary_accuracy: 0.7267 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00
Epoch 6/10
600/600 [==============================] - 0s 108us/step - loss: 0.5853 - binary_accuracy: 0.7283 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - val_loss: 0.5864 - val_binary_accuracy: 0.7267 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00
Epoch 7/10
600/600 [==============================] - 0s 126us/step - loss: 0.5851 - binary_accuracy: 0.7283 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - val_loss: 0.5863 - val_binary_accuracy: 0.7267 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00
Epoch 8/10
600/600 [==============================] - 0s 117us/step - loss: 0.5853 - binary_accuracy: 0.7283 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - val_loss: 0.5862 - val_binary_accuracy: 0.7267 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00
Epoch 9/10
600/600 [==============================] - 0s 158us/step - loss: 0.5854 - binary_accuracy: 0.7283 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - val_loss: 0.5862 - val_binary_accuracy: 0.7267 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00
Epoch 10/10
600/600 [==============================] - 0s 112us/step - loss: 0.5848 - binary_accuracy: 0.7283 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - val_loss: 0.5860 - val_binary_accuracy: 0.7267 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00
[root |INFO|build_network.py:87] Load trained model weight at ./example_result/weight_1.h5
[root |INFO|build_network.py:147] Training end with time 2.74385929107666!
[root |INFO|build_network.py:83] Saved trained model weight at ./example_result/weight_1.h5
[root |DEBUG|deepbiome.py:185] Save weight at ./example_result/weight_1.h5
[root |DEBUG|deepbiome.py:188] Save history at ./example_result/hist_1.json
[root |INFO|build_network.py:173] Evaluation start!
750/750 [==============================] - 0s 5us/step
[root |INFO|build_network.py:178] Evaluation end with time 0.01107335090637207!
[root |INFO|build_network.py:179] Evaluation: [0.5847012400627136, 0.7279999852180481, 1.0, 0.0, 0.0]
[root |INFO|build_network.py:173] Evaluation start!
250/250 [==============================] - 0s 10us/step
[root |INFO|build_network.py:178] Evaluation end with time 0.009724140167236328!
[root |INFO|build_network.py:179] Evaluation: [0.617382287979126, 0.6959999799728394, 1.0, 0.0, 0.0]
[root |INFO|deepbiome.py:199] Compute time : 3.649474859237671
[root |INFO|deepbiome.py:200] 2 fold computing end!---------------------------------------------
[root |INFO|deepbiome.py:153] -------3 simulation start!----------------------------------
[root |INFO|readers.py:58] -----------------------------------------------------------------------
[root |INFO|readers.py:59] Construct Dataset
[root |INFO|readers.py:60] -----------------------------------------------------------------------
[root |INFO|readers.py:61] Load data
[root |INFO|deepbiome.py:164] -----------------------------------------------------------------
[root |INFO|deepbiome.py:165] Build network for 3 simulation
[root |INFO|build_network.py:521] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:522] Read phylogenetic tree information from /DATA/home/muha/github_repos/deepbiome/deepbiome/tests/data/genus48_dic.csv
[root |INFO|build_network.py:528] Phylogenetic tree level list: ['Genus', 'Family', 'Order', 'Class', 'Phylum']
[root |INFO|build_network.py:529] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:537] Genus: 48
[root |INFO|build_network.py:537] Family: 40
[root |INFO|build_network.py:537] Order: 23
[root |INFO|build_network.py:537] Class: 17
[root |INFO|build_network.py:537] Phylum: 9
[root |INFO|build_network.py:546] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:547] Phylogenetic_tree_dict info: ['Phylum', 'Order', 'Family', 'Class', 'Number', 'Genus']
[root |INFO|build_network.py:548] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:558] Build edge weights between [ Genus, Family]
[root |INFO|build_network.py:558] Build edge weights between [Family, Order]
[root |INFO|build_network.py:558] Build edge weights between [ Order, Class]
[root |INFO|build_network.py:558] Build edge weights between [ Class, Phylum]
[root |INFO|build_network.py:571] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:586] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:587] Build network based on phylogenetic tree information
[root |INFO|build_network.py:588] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:670] ------------------------------------------------------------------------------------------
Model: "model_1"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input (InputLayer) (None, 48) 0
_________________________________________________________________
l1_dense (Dense_with_tree) (None, 40) 1960
_________________________________________________________________
l1_activation (Activation) (None, 40) 0
_________________________________________________________________
l2_dense (Dense_with_tree) (None, 23) 943
_________________________________________________________________
l2_activation (Activation) (None, 23) 0
_________________________________________________________________
l3_dense (Dense_with_tree) (None, 17) 408
_________________________________________________________________
l3_activation (Activation) (None, 17) 0
_________________________________________________________________
l4_dense (Dense_with_tree) (None, 9) 162
_________________________________________________________________
l4_activation (Activation) (None, 9) 0
_________________________________________________________________
last_dense_h (Dense) (None, 1) 10
_________________________________________________________________
p_hat (Activation) (None, 1) 0
=================================================================
Total params: 3,483
Trainable params: 3,483
Non-trainable params: 0
_________________________________________________________________
[root |INFO|build_network.py:61] Build Network
[root |INFO|build_network.py:62] Optimizer = adam
[root |INFO|build_network.py:63] Loss = binary_crossentropy
[root |INFO|build_network.py:64] Metrics = binary_accuracy, sensitivity, specificity, gmeasure
[root |INFO|build_network.py:87] Load trained model weight at ./example_result/weight_2.h5
[root |INFO|deepbiome.py:176] -----------------------------------------------------------------
[root |INFO|deepbiome.py:177] 3 fold computing start!----------------------------------
[root |INFO|build_network.py:137] Training start!
Train on 600 samples, validate on 150 samples
Epoch 1/10
600/600 [==============================] - 1s 1ms/step - loss: 0.6220 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - val_loss: 0.6461 - val_binary_accuracy: 0.6533 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00
Epoch 2/10
600/600 [==============================] - 0s 451us/step - loss: 0.6220 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - val_loss: 0.6533 - val_binary_accuracy: 0.6533 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00
Epoch 3/10
600/600 [==============================] - 0s 377us/step - loss: 0.6229 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - val_loss: 0.6460 - val_binary_accuracy: 0.6533 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00
Epoch 4/10
600/600 [==============================] - 0s 134us/step - loss: 0.6208 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - val_loss: 0.6482 - val_binary_accuracy: 0.6533 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00
Epoch 5/10
600/600 [==============================] - 0s 388us/step - loss: 0.6204 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - val_loss: 0.6482 - val_binary_accuracy: 0.6533 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00
Epoch 6/10
600/600 [==============================] - 0s 454us/step - loss: 0.6198 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - val_loss: 0.6467 - val_binary_accuracy: 0.6533 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00
Epoch 7/10
600/600 [==============================] - 0s 149us/step - loss: 0.6202 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - val_loss: 0.6470 - val_binary_accuracy: 0.6533 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00
Epoch 8/10
600/600 [==============================] - 0s 127us/step - loss: 0.6199 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - val_loss: 0.6466 - val_binary_accuracy: 0.6533 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00
Epoch 9/10
600/600 [==============================] - 0s 295us/step - loss: 0.6196 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - val_loss: 0.6454 - val_binary_accuracy: 0.6533 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00
Epoch 10/10
600/600 [==============================] - 0s 529us/step - loss: 0.6201 - binary_accuracy: 0.6883 - sensitivity: 1.0000 - specificity: 0.0000e+00 - gmeasure: 0.0000e+00 - val_loss: 0.6478 - val_binary_accuracy: 0.6533 - val_sensitivity: 1.0000 - val_specificity: 0.0000e+00 - val_gmeasure: 0.0000e+00
[root |INFO|build_network.py:87] Load trained model weight at ./example_result/weight_2.h5
[root |INFO|build_network.py:147] Training end with time 4.314647912979126!
[root |INFO|build_network.py:83] Saved trained model weight at ./example_result/weight_2.h5
[root |DEBUG|deepbiome.py:185] Save weight at ./example_result/weight_2.h5
[root |DEBUG|deepbiome.py:188] Save history at ./example_result/hist_2.json
[root |INFO|build_network.py:173] Evaluation start!
750/750 [==============================] - 0s 3us/step
[root |INFO|build_network.py:178] Evaluation end with time 0.009068727493286133!
[root |INFO|build_network.py:179] Evaluation: [0.6245588660240173, 0.6813333630561829, 1.0, 0.0, 0.0]
[root |INFO|build_network.py:173] Evaluation start!
250/250 [==============================] - 0s 11us/step
[root |INFO|build_network.py:178] Evaluation end with time 0.008772611618041992!
[root |INFO|build_network.py:179] Evaluation: [0.6026607751846313, 0.7120000123977661, 1.0, 0.0, 0.0]
[root |INFO|deepbiome.py:199] Compute time : 4.939546346664429
[root |INFO|deepbiome.py:200] 3 fold computing end!---------------------------------------------
[root |INFO|deepbiome.py:211] -----------------------------------------------------------------
[root |INFO|deepbiome.py:212] Train Evaluation : ['loss' 'binary_accuracy' 'sensitivity' 'specificity' 'gmeasure']
[root |INFO|deepbiome.py:213] mean : [0.570 0.720 0.984 0.099 0.177]
[root |INFO|deepbiome.py:214] std : [0.052 0.028 0.023 0.140 0.251]
[root |INFO|deepbiome.py:215] -----------------------------------------------------------------
[root |INFO|deepbiome.py:216] Test Evaluation : ['loss' 'binary_accuracy' 'sensitivity' 'specificity' 'gmeasure']
[root |INFO|deepbiome.py:217] mean : [0.573 0.719 0.986 0.103 0.181]
[root |INFO|deepbiome.py:218] std : [0.052 0.022 0.020 0.145 0.256]
[root |INFO|deepbiome.py:219] -----------------------------------------------------------------
[root |INFO|deepbiome.py:230] Total Computing Ended
[root |INFO|deepbiome.py:231] -----------------------------------------------------------------
Let’s check the history plot again.
[29]:
with open('./%s/hist_0.json' % path_info['model_info']['model_dir'], 'r') as f:
history = json.load(f)
plt.plot(history['val_loss'], label='Validation')
plt.plot(history['loss'], label='Training')
plt.legend()
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.show()
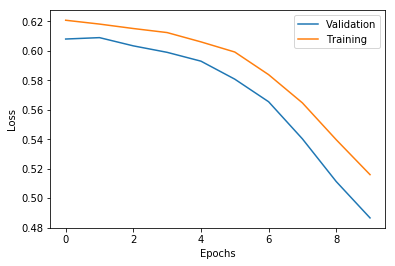
6. Load the pre-trained network for testing¶
To test the trained model, we can use the deepbiome_test function.
If you use the index file (idx_path), this function provides the evaluation using the test index (index set not included in the index file) for each fold. If not, this function provides the evaluation using the whole samples.
If number_of_fold is set to k, the function will test the model only with first k repetitions.
We can use the testing metrics different with the training. In the example below, we additionally used AUC metric.
[30]:
test_network_info = {
'architecture_info': {
'batch_normalization': 'False',
'drop_out': '0',
'weight_initial': 'glorot_uniform',
'weight_l1_penalty':'0.',
'weight_decay': 'phylogenetic_tree',
},
'model_info': {
'lr': '0.01',
'decay': '0.001',
'loss': 'binary_crossentropy',
'metrics': 'binary_accuracy, sensitivity, specificity, gmeasure, auc',
'taxa_selection_metrics': 'sensitivity, specificity, gmeasure, accuracy',
'network_class': 'DeepBiomeNetwork',
'optimizer': 'adam',
'reader_class': 'MicroBiomeClassificationReader',
'normalizer': 'normalize_minmax',
},
'test_info': {
'batch_size': 'None'
}
}
[31]:
test_path_info = {
'data_info': {
'count_list_path': resource_filename('deepbiome', 'tests/data/gcount_list.csv'),
'count_path': resource_filename('deepbiome', 'tests/data/count'),
'data_path': resource_filename('deepbiome', 'tests/data'),
'idx_path': resource_filename('deepbiome', 'tests/data/classification_idx.csv'),
'tree_info_path': resource_filename('deepbiome', 'tests/data/genus48_dic.csv'),
'x_path': '',
'y_path': 'classification_y.csv'
},
'model_info': {
'evaluation': 'eval.npy',
'model_dir': './example_result/',
'weight': 'weight.h5'
}
}
[32]:
evaluation = deepbiome.deepbiome_test(log, test_network_info, test_path_info, number_of_fold=None)
[root |INFO|deepbiome.py:293] -----------------------------------------------------------------
[root |INFO|deepbiome.py:325] Test Evaluation : ['loss' 'binary_accuracy' 'sensitivity' 'specificity' 'gmeasure' 'auc']
[root |INFO|deepbiome.py:327] -------1 fold test start!----------------------------------
[root |INFO|readers.py:58] -----------------------------------------------------------------------
[root |INFO|readers.py:59] Construct Dataset
[root |INFO|readers.py:60] -----------------------------------------------------------------------
[root |INFO|readers.py:61] Load data
[root |INFO|deepbiome.py:338] -----------------------------------------------------------------
[root |INFO|deepbiome.py:339] Build network for 1 fold testing
[root |INFO|build_network.py:521] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:522] Read phylogenetic tree information from /DATA/home/muha/github_repos/deepbiome/deepbiome/tests/data/genus48_dic.csv
[root |INFO|build_network.py:528] Phylogenetic tree level list: ['Genus', 'Family', 'Order', 'Class', 'Phylum']
[root |INFO|build_network.py:529] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:537] Genus: 48
[root |INFO|build_network.py:537] Family: 40
[root |INFO|build_network.py:537] Order: 23
[root |INFO|build_network.py:537] Class: 17
[root |INFO|build_network.py:537] Phylum: 9
[root |INFO|build_network.py:546] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:547] Phylogenetic_tree_dict info: ['Phylum', 'Order', 'Family', 'Class', 'Number', 'Genus']
[root |INFO|build_network.py:548] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:558] Build edge weights between [ Genus, Family]
[root |INFO|build_network.py:558] Build edge weights between [Family, Order]
[root |INFO|build_network.py:558] Build edge weights between [ Order, Class]
[root |INFO|build_network.py:558] Build edge weights between [ Class, Phylum]
[root |INFO|build_network.py:571] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:586] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:587] Build network based on phylogenetic tree information
[root |INFO|build_network.py:588] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:670] ------------------------------------------------------------------------------------------
Model: "model_1"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input (InputLayer) (None, 48) 0
_________________________________________________________________
l1_dense (Dense_with_tree) (None, 40) 1960
_________________________________________________________________
l1_activation (Activation) (None, 40) 0
_________________________________________________________________
l2_dense (Dense_with_tree) (None, 23) 943
_________________________________________________________________
l2_activation (Activation) (None, 23) 0
_________________________________________________________________
l3_dense (Dense_with_tree) (None, 17) 408
_________________________________________________________________
l3_activation (Activation) (None, 17) 0
_________________________________________________________________
l4_dense (Dense_with_tree) (None, 9) 162
_________________________________________________________________
l4_activation (Activation) (None, 9) 0
_________________________________________________________________
last_dense_h (Dense) (None, 1) 10
_________________________________________________________________
p_hat (Activation) (None, 1) 0
=================================================================
Total params: 3,483
Trainable params: 3,483
Non-trainable params: 0
_________________________________________________________________
[root |INFO|build_network.py:61] Build Network
[root |INFO|build_network.py:62] Optimizer = adam
[root |INFO|build_network.py:63] Loss = binary_crossentropy
[root |INFO|build_network.py:64] Metrics = binary_accuracy, sensitivity, specificity, gmeasure, auc
[root |INFO|build_network.py:87] Load trained model weight at ./example_result/weight_0.h5
[root |INFO|deepbiome.py:350] -----------------------------------------------------------------
[root |INFO|deepbiome.py:351] 1 fold computing start!----------------------------------
[root |INFO|build_network.py:173] Evaluation start!
250/250 [==============================] - 0s 352us/step
[root |INFO|build_network.py:178] Evaluation end with time 0.2729969024658203!
[root |INFO|build_network.py:179] Evaluation: [0.5000216364860535, 0.7480000257492065, 0.9585798978805542, 0.3086419701576233, 0.543928325176239, 0.8766162395477295]
[root |INFO|deepbiome.py:356]
[root |INFO|deepbiome.py:357] Compute time : 0.9056384563446045
[root |INFO|deepbiome.py:358] 1 fold computing end!---------------------------------------------
[root |INFO|deepbiome.py:327] -------2 fold test start!----------------------------------
[root |INFO|readers.py:58] -----------------------------------------------------------------------
[root |INFO|readers.py:59] Construct Dataset
[root |INFO|readers.py:60] -----------------------------------------------------------------------
[root |INFO|readers.py:61] Load data
[root |INFO|deepbiome.py:338] -----------------------------------------------------------------
[root |INFO|deepbiome.py:339] Build network for 2 fold testing
[root |INFO|build_network.py:521] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:522] Read phylogenetic tree information from /DATA/home/muha/github_repos/deepbiome/deepbiome/tests/data/genus48_dic.csv
[root |INFO|build_network.py:528] Phylogenetic tree level list: ['Genus', 'Family', 'Order', 'Class', 'Phylum']
[root |INFO|build_network.py:529] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:537] Genus: 48
[root |INFO|build_network.py:537] Family: 40
[root |INFO|build_network.py:537] Order: 23
[root |INFO|build_network.py:537] Class: 17
[root |INFO|build_network.py:537] Phylum: 9
[root |INFO|build_network.py:546] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:547] Phylogenetic_tree_dict info: ['Phylum', 'Order', 'Family', 'Class', 'Number', 'Genus']
[root |INFO|build_network.py:548] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:558] Build edge weights between [ Genus, Family]
[root |INFO|build_network.py:558] Build edge weights between [Family, Order]
[root |INFO|build_network.py:558] Build edge weights between [ Order, Class]
[root |INFO|build_network.py:558] Build edge weights between [ Class, Phylum]
[root |INFO|build_network.py:571] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:586] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:587] Build network based on phylogenetic tree information
[root |INFO|build_network.py:588] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:670] ------------------------------------------------------------------------------------------
Model: "model_1"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input (InputLayer) (None, 48) 0
_________________________________________________________________
l1_dense (Dense_with_tree) (None, 40) 1960
_________________________________________________________________
l1_activation (Activation) (None, 40) 0
_________________________________________________________________
l2_dense (Dense_with_tree) (None, 23) 943
_________________________________________________________________
l2_activation (Activation) (None, 23) 0
_________________________________________________________________
l3_dense (Dense_with_tree) (None, 17) 408
_________________________________________________________________
l3_activation (Activation) (None, 17) 0
_________________________________________________________________
l4_dense (Dense_with_tree) (None, 9) 162
_________________________________________________________________
l4_activation (Activation) (None, 9) 0
_________________________________________________________________
last_dense_h (Dense) (None, 1) 10
_________________________________________________________________
p_hat (Activation) (None, 1) 0
=================================================================
Total params: 3,483
Trainable params: 3,483
Non-trainable params: 0
_________________________________________________________________
[root |INFO|build_network.py:61] Build Network
[root |INFO|build_network.py:62] Optimizer = adam
[root |INFO|build_network.py:63] Loss = binary_crossentropy
[root |INFO|build_network.py:64] Metrics = binary_accuracy, sensitivity, specificity, gmeasure, auc
[root |INFO|build_network.py:87] Load trained model weight at ./example_result/weight_1.h5
[root |INFO|deepbiome.py:350] -----------------------------------------------------------------
[root |INFO|deepbiome.py:351] 2 fold computing start!----------------------------------
[root |INFO|build_network.py:173] Evaluation start!
250/250 [==============================] - 0s 353us/step
[root |INFO|build_network.py:178] Evaluation end with time 0.25853705406188965!
[root |INFO|build_network.py:179] Evaluation: [0.617382287979126, 0.6959999799728394, 1.0, 0.0, 0.0, 0.8632410764694214]
[root |INFO|deepbiome.py:356]
[root |INFO|deepbiome.py:357] Compute time : 0.8716459274291992
[root |INFO|deepbiome.py:358] 2 fold computing end!---------------------------------------------
[root |INFO|deepbiome.py:327] -------3 fold test start!----------------------------------
[root |INFO|readers.py:58] -----------------------------------------------------------------------
[root |INFO|readers.py:59] Construct Dataset
[root |INFO|readers.py:60] -----------------------------------------------------------------------
[root |INFO|readers.py:61] Load data
[root |INFO|deepbiome.py:338] -----------------------------------------------------------------
[root |INFO|deepbiome.py:339] Build network for 3 fold testing
[root |INFO|build_network.py:521] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:522] Read phylogenetic tree information from /DATA/home/muha/github_repos/deepbiome/deepbiome/tests/data/genus48_dic.csv
[root |INFO|build_network.py:528] Phylogenetic tree level list: ['Genus', 'Family', 'Order', 'Class', 'Phylum']
[root |INFO|build_network.py:529] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:537] Genus: 48
[root |INFO|build_network.py:537] Family: 40
[root |INFO|build_network.py:537] Order: 23
[root |INFO|build_network.py:537] Class: 17
[root |INFO|build_network.py:537] Phylum: 9
[root |INFO|build_network.py:546] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:547] Phylogenetic_tree_dict info: ['Phylum', 'Order', 'Family', 'Class', 'Number', 'Genus']
[root |INFO|build_network.py:548] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:558] Build edge weights between [ Genus, Family]
[root |INFO|build_network.py:558] Build edge weights between [Family, Order]
[root |INFO|build_network.py:558] Build edge weights between [ Order, Class]
[root |INFO|build_network.py:558] Build edge weights between [ Class, Phylum]
[root |INFO|build_network.py:571] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:586] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:587] Build network based on phylogenetic tree information
[root |INFO|build_network.py:588] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:670] ------------------------------------------------------------------------------------------
Model: "model_1"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input (InputLayer) (None, 48) 0
_________________________________________________________________
l1_dense (Dense_with_tree) (None, 40) 1960
_________________________________________________________________
l1_activation (Activation) (None, 40) 0
_________________________________________________________________
l2_dense (Dense_with_tree) (None, 23) 943
_________________________________________________________________
l2_activation (Activation) (None, 23) 0
_________________________________________________________________
l3_dense (Dense_with_tree) (None, 17) 408
_________________________________________________________________
l3_activation (Activation) (None, 17) 0
_________________________________________________________________
l4_dense (Dense_with_tree) (None, 9) 162
_________________________________________________________________
l4_activation (Activation) (None, 9) 0
_________________________________________________________________
last_dense_h (Dense) (None, 1) 10
_________________________________________________________________
p_hat (Activation) (None, 1) 0
=================================================================
Total params: 3,483
Trainable params: 3,483
Non-trainable params: 0
_________________________________________________________________
[root |INFO|build_network.py:61] Build Network
[root |INFO|build_network.py:62] Optimizer = adam
[root |INFO|build_network.py:63] Loss = binary_crossentropy
[root |INFO|build_network.py:64] Metrics = binary_accuracy, sensitivity, specificity, gmeasure, auc
[root |INFO|build_network.py:87] Load trained model weight at ./example_result/weight_2.h5
[root |INFO|deepbiome.py:350] -----------------------------------------------------------------
[root |INFO|deepbiome.py:351] 3 fold computing start!----------------------------------
[root |INFO|build_network.py:173] Evaluation start!
250/250 [==============================] - 0s 381us/step
[root |INFO|build_network.py:178] Evaluation end with time 0.28519654273986816!
[root |INFO|build_network.py:179] Evaluation: [0.6026607751846313, 0.7120000123977661, 1.0, 0.0, 0.0, 0.5095973610877991]
[root |INFO|deepbiome.py:356]
[root |INFO|deepbiome.py:357] Compute time : 0.8778982162475586
[root |INFO|deepbiome.py:358] 3 fold computing end!---------------------------------------------
[root |INFO|deepbiome.py:367] -----------------------------------------------------------------
[root |INFO|deepbiome.py:368] Test Evaluation : ['loss' 'binary_accuracy' 'sensitivity' 'specificity' 'gmeasure' 'auc']
[root |INFO|deepbiome.py:369] mean : [0.573 0.719 0.986 0.103 0.181 0.750]
[root |INFO|deepbiome.py:370] std : [0.052 0.022 0.020 0.145 0.256 0.170]
[root |INFO|deepbiome.py:371] -----------------------------------------------------------------
[root |INFO|deepbiome.py:372] Total Computing Ended
[root |INFO|deepbiome.py:373] -----------------------------------------------------------------
This function provides the evaluation result as a numpy array with a shape of (number of repetition, number of evaluation measures).
[33]:
print(' %s' % ''.join(['%16s'%'loss']+ ['%16s'%s.strip() for s in network_info['model_info']['metrics'].split(',')]))
print('Mean: %s' % ''.join(['%16.3f'%v for v in np.mean(evaluation, axis=0)]))
print('Std : %s' % ''.join(['%16.3f'%v for v in np.std(evaluation, axis=0)]))
loss binary_accuracy sensitivity specificity gmeasure auc
Mean: 0.573 0.719 0.986 0.103 0.181 0.750
Std : 0.052 0.022 0.020 0.145 0.256 0.170
7. Load the pre-trained network for prediction¶
If you want to predict using the pre-trained model, you can use the deepbiome_prediction function. If number_of_fold is setted as k, the function will predict only with first k repetitions sample’s outputs.
If change_weight_for_each_fold is set as False, the function will predict the output of every repetition by same weight from the given path. If change_weight_for_each_fold is set as True, the function will predict the output of by each repetition weight.
If ‘get_y=True’, the function will provide a list of tuples (prediction, true output) as a output with the shape of (n_samples, 2, n_classes). If ‘get_y=False’, the function will provide predictions only. The output will have the shape of (n_samples, n_classes).
7.1 Prediction with fixed weight¶
If we want to predict new data from one pre-trained model, we can use the option below. We fixed the weight weight_0.h5 for predicting the whole samples (without using index file).
[34]:
prediction_network_info = {
'architecture_info': {
'batch_normalization': 'False',
'drop_out': '0',
'weight_initial': 'glorot_uniform',
'weight_l1_penalty':'0.',
'weight_decay': 'phylogenetic_tree',
},
'model_info': {
'decay': '0.001',
'loss': 'binary_crossentropy',
'lr': '0.01',
'metrics': 'binary_accuracy, sensitivity, specificity, gmeasure, auc',
'network_class': 'DeepBiomeNetwork',
'normalizer': 'normalize_minmax',
'optimizer': 'adam',
'reader_class': 'MicroBiomeClassificationReader',
'taxa_selection_metrics': 'sensitivity, specificity, gmeasure, accuracy',
},
'test_info': {
'batch_size': 'None'
}
}
[35]:
prediction_path_info = {
'data_info': {
'count_list_path': resource_filename('deepbiome', 'tests/data/gcount_list.csv'),
'count_path': resource_filename('deepbiome', 'tests/data/count'),
'data_path': resource_filename('deepbiome', 'tests/data'),
'tree_info_path': resource_filename('deepbiome', 'tests/data/genus48_dic.csv'),
'x_path': '',
},
'model_info': {
'model_dir': './example_result/',
'weight': 'weight_0.h5'
}
}
[36]:
prediction = deepbiome.deepbiome_prediction(log, prediction_network_info, prediction_path_info,
num_classes = 1, number_of_fold=None)
[root |INFO|deepbiome.py:450] -----------------------------------------------------------------
[root |INFO|deepbiome.py:480] -------1 th repeatition prediction start!----------------------------------
[root |INFO|readers.py:58] -----------------------------------------------------------------------
[root |INFO|readers.py:59] Construct Dataset
[root |INFO|readers.py:60] -----------------------------------------------------------------------
[root |INFO|readers.py:61] Load data
[root |INFO|deepbiome.py:498] -----------------------------------------------------------------
[root |INFO|build_network.py:521] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:522] Read phylogenetic tree information from /DATA/home/muha/github_repos/deepbiome/deepbiome/tests/data/genus48_dic.csv
[root |INFO|build_network.py:528] Phylogenetic tree level list: ['Genus', 'Family', 'Order', 'Class', 'Phylum']
[root |INFO|build_network.py:529] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:537] Genus: 48
[root |INFO|build_network.py:537] Family: 40
[root |INFO|build_network.py:537] Order: 23
[root |INFO|build_network.py:537] Class: 17
[root |INFO|build_network.py:537] Phylum: 9
[root |INFO|build_network.py:546] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:547] Phylogenetic_tree_dict info: ['Phylum', 'Order', 'Family', 'Class', 'Number', 'Genus']
[root |INFO|build_network.py:548] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:558] Build edge weights between [ Genus, Family]
[root |INFO|build_network.py:558] Build edge weights between [Family, Order]
[root |INFO|build_network.py:558] Build edge weights between [ Order, Class]
[root |INFO|build_network.py:558] Build edge weights between [ Class, Phylum]
[root |INFO|build_network.py:571] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:586] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:587] Build network based on phylogenetic tree information
[root |INFO|build_network.py:588] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:670] ------------------------------------------------------------------------------------------
Model: "model_1"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input (InputLayer) (None, 48) 0
_________________________________________________________________
l1_dense (Dense_with_tree) (None, 40) 1960
_________________________________________________________________
l1_activation (Activation) (None, 40) 0
_________________________________________________________________
l2_dense (Dense_with_tree) (None, 23) 943
_________________________________________________________________
l2_activation (Activation) (None, 23) 0
_________________________________________________________________
l3_dense (Dense_with_tree) (None, 17) 408
_________________________________________________________________
l3_activation (Activation) (None, 17) 0
_________________________________________________________________
l4_dense (Dense_with_tree) (None, 9) 162
_________________________________________________________________
l4_activation (Activation) (None, 9) 0
_________________________________________________________________
last_dense_h (Dense) (None, 1) 10
_________________________________________________________________
p_hat (Activation) (None, 1) 0
=================================================================
Total params: 3,483
Trainable params: 3,483
Non-trainable params: 0
_________________________________________________________________
[root |INFO|build_network.py:61] Build Network
[root |INFO|build_network.py:62] Optimizer = adam
[root |INFO|build_network.py:63] Loss = binary_crossentropy
[root |INFO|build_network.py:64] Metrics = binary_accuracy, sensitivity, specificity, gmeasure, auc
[root |INFO|build_network.py:87] Load trained model weight at ./example_result/weight_0.h5
[root |INFO|deepbiome.py:509] -----------------------------------------------------------------
[root |INFO|build_network.py:193] Prediction start!
1000/1000 [==============================] - 0s 49us/step
[root |INFO|build_network.py:198] Prediction end with time 0.052338600158691406!
[root |INFO|deepbiome.py:513] Compute time : 0.6934528350830078
[root |INFO|deepbiome.py:514] 1 fold computing end!---------------------------------------------
[root |INFO|deepbiome.py:519] Total Computing Ended
[root |INFO|deepbiome.py:520] -----------------------------------------------------------------
[37]:
prediction.shape
[37]:
(1, 1000, 1)
[38]:
prediction[0,:10]
[38]:
array([[0.779],
[0.771],
[0.620],
[0.501],
[0.863],
[0.622],
[0.630],
[0.496],
[0.822],
[0.843]], dtype=float32)
7.2 Prediction with each fold weight¶
If we want to predict the test outputs from each repetitions, we can use the option belows.
The example below shows how to predict the 5 repetition outputs. We set idx_path for using the index file classification_idx.csv to predict only the test set for each repetition.
[39]:
prediction_network_info = {
'architecture_info': {
'batch_normalization': 'False',
'drop_out': '0',
'weight_initial': 'glorot_uniform',
'weight_l1_penalty':'0.',
'weight_decay': 'phylogenetic_tree',
},
'model_info': {
'decay': '0.001',
'loss': 'binary_crossentropy',
'lr': '0.01',
'metrics': 'binary_accuracy, sensitivity, specificity, gmeasure, auc',
'network_class': 'DeepBiomeNetwork',
'normalizer': 'normalize_minmax',
'optimizer': 'adam',
'reader_class': 'MicroBiomeClassificationReader',
'taxa_selection_metrics': 'sensitivity, specificity, gmeasure, accuracy',
},
'test_info': {
'batch_size': 'None'
}
}
[40]:
prediction_path_info = {
'data_info': {
'count_list_path': resource_filename('deepbiome', 'tests/data/gcount_list.csv'),
'count_path': resource_filename('deepbiome', 'tests/data/count'),
'data_path': resource_filename('deepbiome', 'tests/data'),
'idx_path': resource_filename('deepbiome', 'tests/data/classification_idx.csv'),
'tree_info_path': resource_filename('deepbiome', 'tests/data/genus48_dic.csv'),
'x_path': '',
'y_path': 'classification_y.csv'
},
'model_info': {
'model_dir': './example_result/',
'weight': 'weight.h5'
}
}
To predict the CV outputs from each fold, we set change_weight_for_each_fold = True. Also, we set get_y=True to get the paired output of each prediction too.
[41]:
prediction = deepbiome.deepbiome_prediction(log, prediction_network_info, prediction_path_info,
num_classes = 1, number_of_fold=None,
change_weight_for_each_fold = True,
get_y=True)
[root |INFO|deepbiome.py:450] -----------------------------------------------------------------
[root |INFO|deepbiome.py:480] -------1 th repeatition prediction start!----------------------------------
[root |INFO|readers.py:58] -----------------------------------------------------------------------
[root |INFO|readers.py:59] Construct Dataset
[root |INFO|readers.py:60] -----------------------------------------------------------------------
[root |INFO|readers.py:61] Load data
[root |INFO|deepbiome.py:498] -----------------------------------------------------------------
[root |INFO|build_network.py:521] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:522] Read phylogenetic tree information from /DATA/home/muha/github_repos/deepbiome/deepbiome/tests/data/genus48_dic.csv
[root |INFO|build_network.py:528] Phylogenetic tree level list: ['Genus', 'Family', 'Order', 'Class', 'Phylum']
[root |INFO|build_network.py:529] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:537] Genus: 48
[root |INFO|build_network.py:537] Family: 40
[root |INFO|build_network.py:537] Order: 23
[root |INFO|build_network.py:537] Class: 17
[root |INFO|build_network.py:537] Phylum: 9
[root |INFO|build_network.py:546] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:547] Phylogenetic_tree_dict info: ['Phylum', 'Order', 'Family', 'Class', 'Number', 'Genus']
[root |INFO|build_network.py:548] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:558] Build edge weights between [ Genus, Family]
[root |INFO|build_network.py:558] Build edge weights between [Family, Order]
[root |INFO|build_network.py:558] Build edge weights between [ Order, Class]
[root |INFO|build_network.py:558] Build edge weights between [ Class, Phylum]
[root |INFO|build_network.py:571] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:586] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:587] Build network based on phylogenetic tree information
[root |INFO|build_network.py:588] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:670] ------------------------------------------------------------------------------------------
Model: "model_1"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input (InputLayer) (None, 48) 0
_________________________________________________________________
l1_dense (Dense_with_tree) (None, 40) 1960
_________________________________________________________________
l1_activation (Activation) (None, 40) 0
_________________________________________________________________
l2_dense (Dense_with_tree) (None, 23) 943
_________________________________________________________________
l2_activation (Activation) (None, 23) 0
_________________________________________________________________
l3_dense (Dense_with_tree) (None, 17) 408
_________________________________________________________________
l3_activation (Activation) (None, 17) 0
_________________________________________________________________
l4_dense (Dense_with_tree) (None, 9) 162
_________________________________________________________________
l4_activation (Activation) (None, 9) 0
_________________________________________________________________
last_dense_h (Dense) (None, 1) 10
_________________________________________________________________
p_hat (Activation) (None, 1) 0
=================================================================
Total params: 3,483
Trainable params: 3,483
Non-trainable params: 0
_________________________________________________________________
[root |INFO|build_network.py:61] Build Network
[root |INFO|build_network.py:62] Optimizer = adam
[root |INFO|build_network.py:63] Loss = binary_crossentropy
[root |INFO|build_network.py:64] Metrics = binary_accuracy, sensitivity, specificity, gmeasure, auc
[root |INFO|build_network.py:87] Load trained model weight at ./example_result/weight_0.h5
[root |INFO|deepbiome.py:509] -----------------------------------------------------------------
[root |INFO|build_network.py:193] Prediction start!
250/250 [==============================] - 0s 136us/step
[root |INFO|build_network.py:198] Prediction end with time 0.03711438179016113!
[root |INFO|deepbiome.py:513] Compute time : 0.6130969524383545
[root |INFO|deepbiome.py:514] 1 fold computing end!---------------------------------------------
[root |INFO|deepbiome.py:480] -------2 th repeatition prediction start!----------------------------------
[root |INFO|readers.py:58] -----------------------------------------------------------------------
[root |INFO|readers.py:59] Construct Dataset
[root |INFO|readers.py:60] -----------------------------------------------------------------------
[root |INFO|readers.py:61] Load data
[root |INFO|deepbiome.py:498] -----------------------------------------------------------------
[root |INFO|build_network.py:521] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:522] Read phylogenetic tree information from /DATA/home/muha/github_repos/deepbiome/deepbiome/tests/data/genus48_dic.csv
[root |INFO|build_network.py:528] Phylogenetic tree level list: ['Genus', 'Family', 'Order', 'Class', 'Phylum']
[root |INFO|build_network.py:529] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:537] Genus: 48
[root |INFO|build_network.py:537] Family: 40
[root |INFO|build_network.py:537] Order: 23
[root |INFO|build_network.py:537] Class: 17
[root |INFO|build_network.py:537] Phylum: 9
[root |INFO|build_network.py:546] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:547] Phylogenetic_tree_dict info: ['Phylum', 'Order', 'Family', 'Class', 'Number', 'Genus']
[root |INFO|build_network.py:548] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:558] Build edge weights between [ Genus, Family]
[root |INFO|build_network.py:558] Build edge weights between [Family, Order]
[root |INFO|build_network.py:558] Build edge weights between [ Order, Class]
[root |INFO|build_network.py:558] Build edge weights between [ Class, Phylum]
[root |INFO|build_network.py:571] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:586] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:587] Build network based on phylogenetic tree information
[root |INFO|build_network.py:588] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:670] ------------------------------------------------------------------------------------------
Model: "model_2"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input (InputLayer) (None, 48) 0
_________________________________________________________________
l1_dense (Dense_with_tree) (None, 40) 1960
_________________________________________________________________
l1_activation (Activation) (None, 40) 0
_________________________________________________________________
l2_dense (Dense_with_tree) (None, 23) 943
_________________________________________________________________
l2_activation (Activation) (None, 23) 0
_________________________________________________________________
l3_dense (Dense_with_tree) (None, 17) 408
_________________________________________________________________
l3_activation (Activation) (None, 17) 0
_________________________________________________________________
l4_dense (Dense_with_tree) (None, 9) 162
_________________________________________________________________
l4_activation (Activation) (None, 9) 0
_________________________________________________________________
last_dense_h (Dense) (None, 1) 10
_________________________________________________________________
p_hat (Activation) (None, 1) 0
=================================================================
Total params: 3,483
Trainable params: 3,483
Non-trainable params: 0
_________________________________________________________________
[root |INFO|build_network.py:61] Build Network
[root |INFO|build_network.py:62] Optimizer = adam
[root |INFO|build_network.py:63] Loss = binary_crossentropy
[root |INFO|build_network.py:64] Metrics = binary_accuracy, sensitivity, specificity, gmeasure, auc
[root |INFO|build_network.py:87] Load trained model weight at ./example_result/weight_1.h5
[root |INFO|deepbiome.py:509] -----------------------------------------------------------------
[root |INFO|build_network.py:193] Prediction start!
250/250 [==============================] - 0s 271us/step
[root |INFO|build_network.py:198] Prediction end with time 0.07141661643981934!
[root |INFO|deepbiome.py:513] Compute time : 0.6672861576080322
[root |INFO|deepbiome.py:514] 2 fold computing end!---------------------------------------------
[root |INFO|deepbiome.py:480] -------3 th repeatition prediction start!----------------------------------
[root |INFO|readers.py:58] -----------------------------------------------------------------------
[root |INFO|readers.py:59] Construct Dataset
[root |INFO|readers.py:60] -----------------------------------------------------------------------
[root |INFO|readers.py:61] Load data
[root |INFO|deepbiome.py:498] -----------------------------------------------------------------
[root |INFO|build_network.py:521] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:522] Read phylogenetic tree information from /DATA/home/muha/github_repos/deepbiome/deepbiome/tests/data/genus48_dic.csv
[root |INFO|build_network.py:528] Phylogenetic tree level list: ['Genus', 'Family', 'Order', 'Class', 'Phylum']
[root |INFO|build_network.py:529] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:537] Genus: 48
[root |INFO|build_network.py:537] Family: 40
[root |INFO|build_network.py:537] Order: 23
[root |INFO|build_network.py:537] Class: 17
[root |INFO|build_network.py:537] Phylum: 9
[root |INFO|build_network.py:546] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:547] Phylogenetic_tree_dict info: ['Phylum', 'Order', 'Family', 'Class', 'Number', 'Genus']
[root |INFO|build_network.py:548] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:558] Build edge weights between [ Genus, Family]
[root |INFO|build_network.py:558] Build edge weights between [Family, Order]
[root |INFO|build_network.py:558] Build edge weights between [ Order, Class]
[root |INFO|build_network.py:558] Build edge weights between [ Class, Phylum]
[root |INFO|build_network.py:571] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:586] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:587] Build network based on phylogenetic tree information
[root |INFO|build_network.py:588] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:670] ------------------------------------------------------------------------------------------
Model: "model_3"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input (InputLayer) (None, 48) 0
_________________________________________________________________
l1_dense (Dense_with_tree) (None, 40) 1960
_________________________________________________________________
l1_activation (Activation) (None, 40) 0
_________________________________________________________________
l2_dense (Dense_with_tree) (None, 23) 943
_________________________________________________________________
l2_activation (Activation) (None, 23) 0
_________________________________________________________________
l3_dense (Dense_with_tree) (None, 17) 408
_________________________________________________________________
l3_activation (Activation) (None, 17) 0
_________________________________________________________________
l4_dense (Dense_with_tree) (None, 9) 162
_________________________________________________________________
l4_activation (Activation) (None, 9) 0
_________________________________________________________________
last_dense_h (Dense) (None, 1) 10
_________________________________________________________________
p_hat (Activation) (None, 1) 0
=================================================================
Total params: 3,483
Trainable params: 3,483
Non-trainable params: 0
_________________________________________________________________
[root |INFO|build_network.py:61] Build Network
[root |INFO|build_network.py:62] Optimizer = adam
[root |INFO|build_network.py:63] Loss = binary_crossentropy
[root |INFO|build_network.py:64] Metrics = binary_accuracy, sensitivity, specificity, gmeasure, auc
[root |INFO|build_network.py:87] Load trained model weight at ./example_result/weight_2.h5
[root |INFO|deepbiome.py:509] -----------------------------------------------------------------
[root |INFO|build_network.py:193] Prediction start!
250/250 [==============================] - 0s 301us/step
[root |INFO|build_network.py:198] Prediction end with time 0.07864809036254883!
[root |INFO|deepbiome.py:513] Compute time : 0.7205522060394287
[root |INFO|deepbiome.py:514] 3 fold computing end!---------------------------------------------
[root |INFO|deepbiome.py:519] Total Computing Ended
[root |INFO|deepbiome.py:520] -----------------------------------------------------------------
We gathered the outputs from each fold.
[42]:
prediction = np.vstack(prediction)
Since we set the option get_y=True, the output has the shape of (n_samples, 2, n_classes). With this options, we can get the predictions of test set and the true output of each predictions.
Now, we can calculate the test performance by the test predictions.
[43]:
predict_output = prediction[:,0]
true_output = prediction[:,1]
log.info('Shape of the predict function ourput: %s' % str(prediction.shape))
log.info('Shape of the prediction: %s' % str(predict_output.shape))
log.info('Shape of the true_output for each prediction: %s' % str(true_output.shape))
[root |INFO|<ipython-input-43-dc5b95be41ff>:4] Shape of the predict function ourput: (750, 2, 1)
[root |INFO|<ipython-input-43-dc5b95be41ff>:5] Shape of the prediction: (750, 1)
[root |INFO|<ipython-input-43-dc5b95be41ff>:6] Shape of the true_output for each prediction: (750, 1)
[44]:
log.info('CV accuracy: %6.3f' % np.mean((predict_output >= 0.5) == true_output))
[root |INFO|<ipython-input-44-ecaee2413087>:1] CV accuracy: 0.719
8. Load trained weight matrix¶
The deepbiome_get_trained_weight function convert the trained weight *.h5 saved from the deepbiome_train to a list of numpy arrays. In this exampe, the list has numpy array of weights from 6 layers. ([genus to family, family to order, order to Class, class to phylum, phylum to output])
[45]:
weight_path = '%s/%s' % (path_info['model_info']['model_dir'], 'weight_0.h5')
trained_weight_list = deepbiome.deepbiome_get_trained_weight(log, network_info, path_info, num_classes=1, weight_path=weight_path)
log.info(len(trained_weight_list))
[root |INFO|readers.py:58] -----------------------------------------------------------------------
[root |INFO|readers.py:59] Construct Dataset
[root |INFO|readers.py:60] -----------------------------------------------------------------------
[root |INFO|readers.py:61] Load data
[root |INFO|build_network.py:521] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:522] Read phylogenetic tree information from /DATA/home/muha/github_repos/deepbiome/deepbiome/tests/data/genus48_dic.csv
[root |INFO|build_network.py:528] Phylogenetic tree level list: ['Genus', 'Family', 'Order', 'Class', 'Phylum']
[root |INFO|build_network.py:529] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:537] Genus: 48
[root |INFO|build_network.py:537] Family: 40
[root |INFO|build_network.py:537] Order: 23
[root |INFO|build_network.py:537] Class: 17
[root |INFO|build_network.py:537] Phylum: 9
[root |INFO|build_network.py:546] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:547] Phylogenetic_tree_dict info: ['Phylum', 'Order', 'Family', 'Class', 'Number', 'Genus']
[root |INFO|build_network.py:548] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:558] Build edge weights between [ Genus, Family]
[root |INFO|build_network.py:558] Build edge weights between [Family, Order]
[root |INFO|build_network.py:558] Build edge weights between [ Order, Class]
[root |INFO|build_network.py:558] Build edge weights between [ Class, Phylum]
[root |INFO|build_network.py:571] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:586] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:587] Build network based on phylogenetic tree information
[root |INFO|build_network.py:588] ------------------------------------------------------------------------------------------
[root |INFO|build_network.py:670] ------------------------------------------------------------------------------------------
Model: "model_1"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input (InputLayer) (None, 48) 0
_________________________________________________________________
l1_dense (Dense_with_tree) (None, 40) 1960
_________________________________________________________________
l1_activation (Activation) (None, 40) 0
_________________________________________________________________
l2_dense (Dense_with_tree) (None, 23) 943
_________________________________________________________________
l2_activation (Activation) (None, 23) 0
_________________________________________________________________
l3_dense (Dense_with_tree) (None, 17) 408
_________________________________________________________________
l3_activation (Activation) (None, 17) 0
_________________________________________________________________
l4_dense (Dense_with_tree) (None, 9) 162
_________________________________________________________________
l4_activation (Activation) (None, 9) 0
_________________________________________________________________
last_dense_h (Dense) (None, 1) 10
_________________________________________________________________
p_hat (Activation) (None, 1) 0
=================================================================
Total params: 3,483
Trainable params: 3,483
Non-trainable params: 0
_________________________________________________________________
[root |INFO|<ipython-input-45-9505ee8dcaa8>:3] 5
First weight between the genus and family layers has the shape of (number of genus = 48, number of family = 40)
[46]:
log.info(trained_weight_list[0].shape)
[root |INFO|<ipython-input-46-c71fa46ab178>:1] (48, 40)
9. Taxa selection performance¶
If we know the true disease path, we can calculate the taxa selection performance by deepbiome_taxa_selection_performance funciton. First, we prepared the true weight list based on the true disease path. For each fold, we prepared 4 weights from the 5 layers ([genus to family, family to order, order to Class, class to phylum]). An example of the list of the true weights from each fold is as follow:
[47]:
true_tree_weight_list = np.load(resource_filename('deepbiome', 'tests/data/true_weight_list.npy'), allow_pickle=True)
log.info(true_tree_weight_list.shape)
[root |INFO|<ipython-input-47-7f16305fbcb7>:2] (5, 4)
The first weight between the genus and family layers for first epoch has the shape below:
[48]:
log.info(true_tree_weight_list[0][0].shape)
[root |INFO|<ipython-input-48-7f1406e7d9a7>:1] (48, 40)
We will calculate the taxa selection performance of the trained weight below:
[49]:
trained_weight_path_list = ['%s/weight_%d.h5' % (path_info['model_info']['model_dir'], i) for i in range(3)]
trained_weight_path_list
[49]:
['./example_result//weight_0.h5',
'./example_result//weight_1.h5',
'./example_result//weight_2.h5']
This is the summary of the taxa selection accuracy of trained weights from each fold.
[50]:
summary = deepbiome.deepbiome_taxa_selection_performance(log, network_info, path_info, num_classes=1,
true_tree_weight_list=true_tree_weight_list,
trained_weight_path_list = trained_weight_path_list)
[root |INFO|readers.py:58] -----------------------------------------------------------------------
[root |INFO|readers.py:59] Construct Dataset
[root |INFO|readers.py:60] -----------------------------------------------------------------------
[root |INFO|readers.py:61] Load data
[root |INFO|build_network.py:87] Load trained model weight at ./example_result//weight_0.h5
[root |INFO|build_network.py:87] Load trained model weight at ./example_result//weight_1.h5
[root |INFO|build_network.py:87] Load trained model weight at ./example_result//weight_2.h5
[51]:
summary
[51]:
| Model | PhyloTree | No. true taxa | No. total taxa | Sensitivity_mean | Sensitivity_std | Specificity_mean | Specificity_std | Gmeasure_mean | Gmeasure_std | Accuracy_mean | Accuracy_std | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | ./example_result/ | Genus | 31 | 48 | 0.946 | 0.055 | 0.983 | 0.009 | 0.964 | 0.026 | 0.983 | 0.008 |
| 1 | Family | 23 | 40 | 0.942 | 0.020 | 0.981 | 0.002 | 0.961 | 0.011 | 0.980 | 0.002 | |
| 2 | Order | 9 | 23 | 0.963 | 0.052 | 0.962 | 0.002 | 0.962 | 0.028 | 0.962 | 0.004 | |
| 3 | Class | 7 | 17 | 0.952 | 0.067 | 0.925 | 0.010 | 0.938 | 0.032 | 0.926 | 0.008 |